null
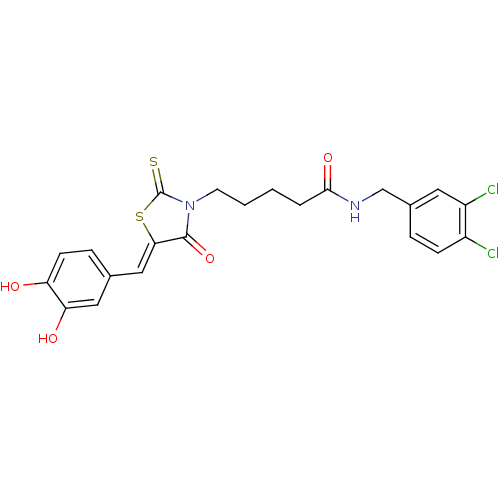
SMILES Oc1ccc(\C=C2/SC(=S)N(CCCCC(=O)NCc3ccc(Cl)c(Cl)c3)C2=O)cc1O
InChI Key InChIKey=QJYBFPPSYZFEAR-ODLFYWEKSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 59099
Found 3 hits for monomerid = 59099
Affinity DataKi: 42nMpH: 7.4Assay Description:All reactions were monitored spectrophotometrically at 340 nm by using initial rates from the first 5% of reaction.More data for this Ligand-Target Pair
Affinity DataKi: 1.00E+4nMpH: 7.4Assay Description:All reactions were monitored spectrophotometrically at 340 nm by using initial rates from the first 5% of reaction.More data for this Ligand-Target Pair
Affinity DataKi: >5.00E+4nMpH: 7.4Assay Description:All reactions were monitored spectrophotometrically at 340 nm by using initial rates from the first 5% of reaction.More data for this Ligand-Target Pair