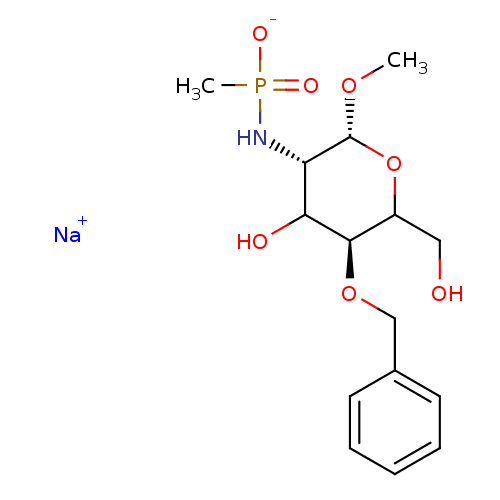
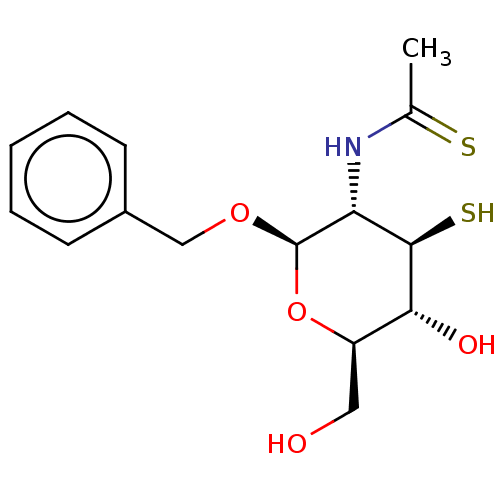
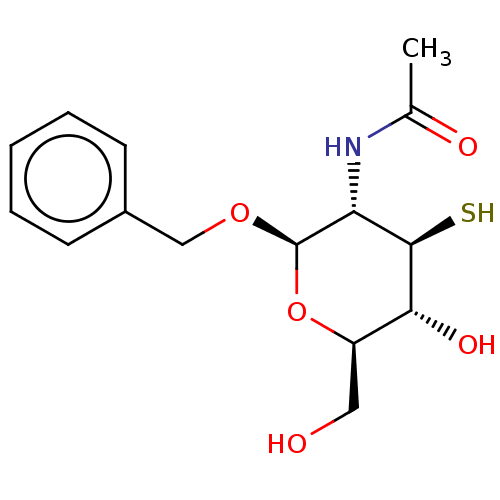
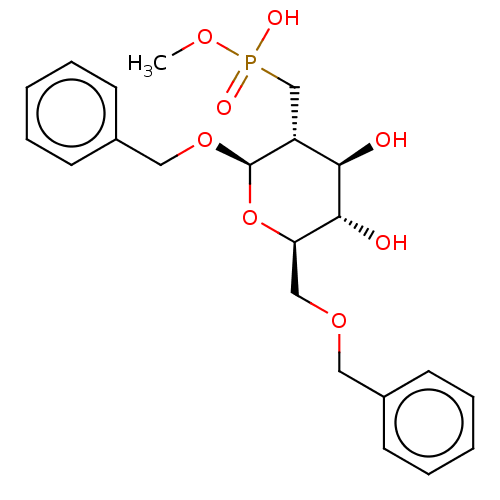
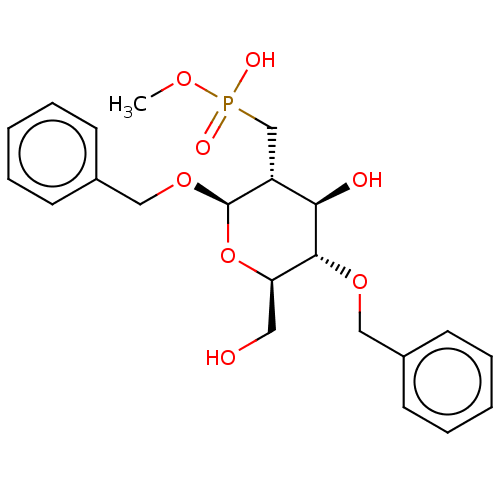
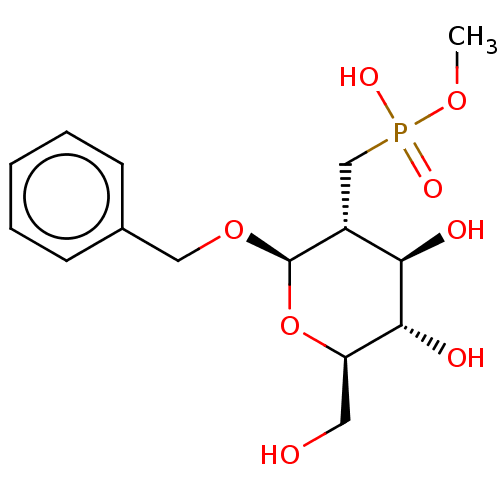
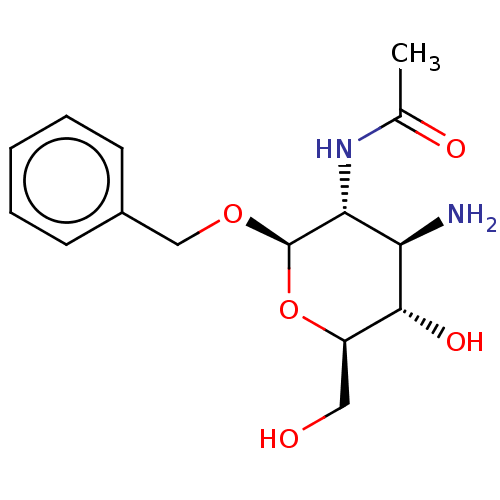
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataKi: >1.00E+6nMpH: 7.5Assay Description:Reactions were carried out in solutions (50 無) containing PgaB (10 然) in HEPES (100 mM, pH 7.5), and varying concentrations of ACC (0.05-10 mM) at ...More data for this Ligand-Target Pair
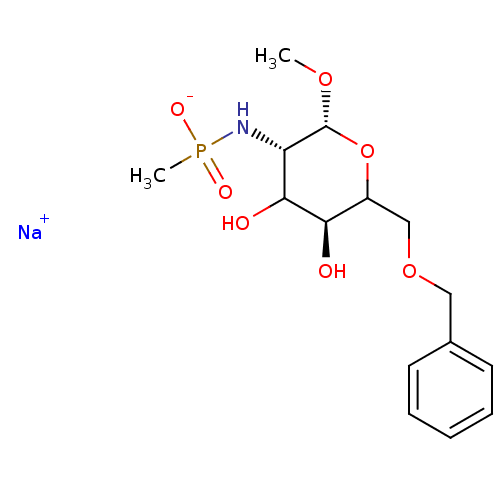
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataKi: >1.00E+6nMpH: 7.5Assay Description:Reactions were carried out in solutions (50 無) containing PgaB (10 然) in HEPES (100 mM, pH 7.5), and varying concentrations of ACC (0.05-10 mM) at ...More data for this Ligand-Target Pair
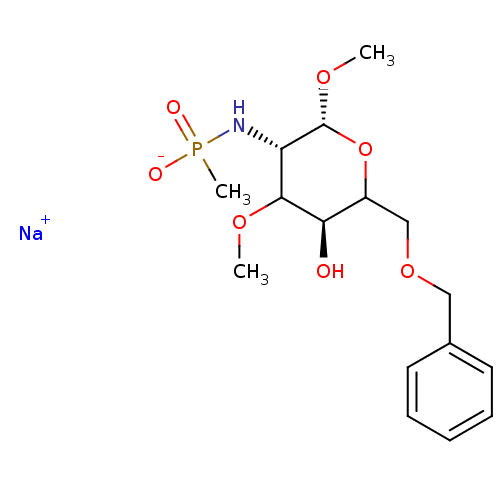
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataKi: >1.00E+6nMpH: 7.5Assay Description:Reactions were carried out in solutions (50 無) containing PgaB (10 然) in HEPES (100 mM, pH 7.5), and varying concentrations of ACC (0.05-10 mM) at ...More data for this Ligand-Target Pair
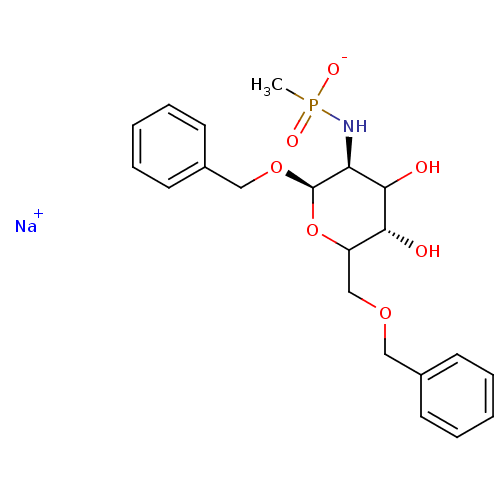
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataKi: >1.00E+6nMpH: 7.5Assay Description:Reactions were carried out in solutions (50 無) containing PgaB (10 然) in HEPES (100 mM, pH 7.5), and varying concentrations of ACC (0.05-10 mM) at ...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataKi: >1.00E+6nMpH: 7.5Assay Description:Reactions were carried out in solutions (50 無) containing PgaB (10 然) in HEPES (100 mM, pH 7.5), and varying concentrations of ACC (0.05-10 mM) at ...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 1.40E+5nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using compoun...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using compoun...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using compoun...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 1.00E+6nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using compoun...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 1.10E+6nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using compoun...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 5.00E+6nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using acetoxy...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 5.00E+6nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using acetoxy...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 5.00E+6nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using acetoxy...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 5.00E+6nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using acetoxy...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 8.00E+6nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using compoun...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 1.00E+7nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using acetoxy...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 1.00E+7nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using acetoxy...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 1.00E+7nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using acetoxy...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 1.00E+7nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using acetoxy...More data for this Ligand-Target Pair
TargetPoly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase(Escherichia coli (Enterobacteria))
University of Toronto
University of Toronto
Affinity DataIC50: 2.00E+7nMAssay Description:Inhibition of Escherichia coli PgaB expressed in Escherichia coli BL21 (DE3) cells transformed with pET28 plasmid coding for PgaB42-655 using acetoxy...More data for this Ligand-Target Pair