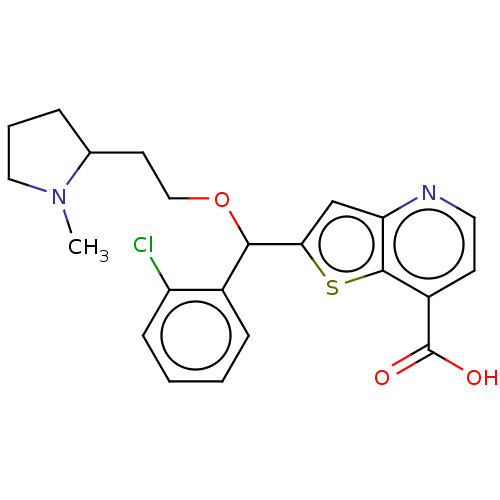
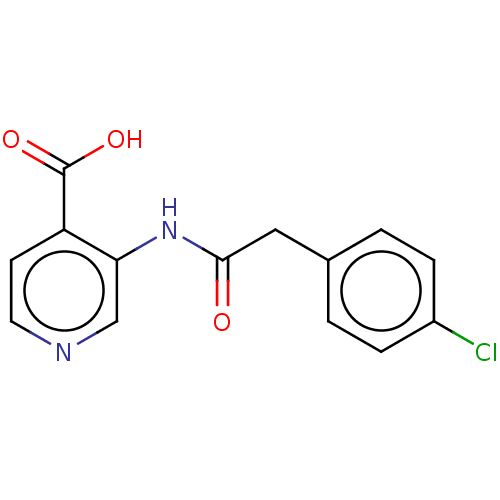
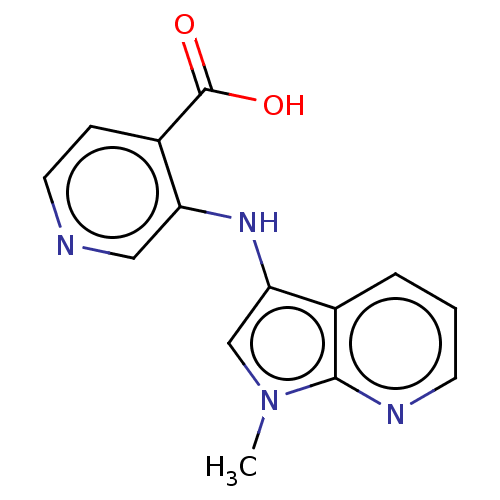
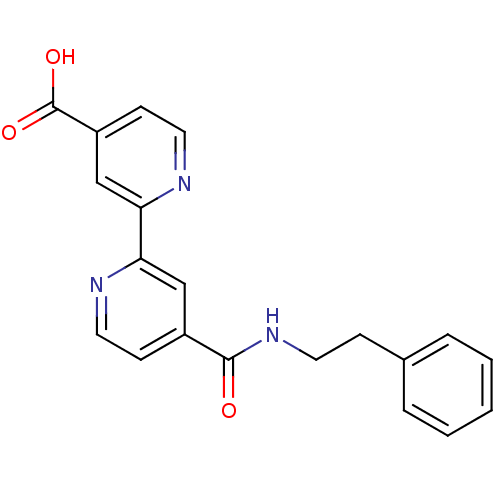
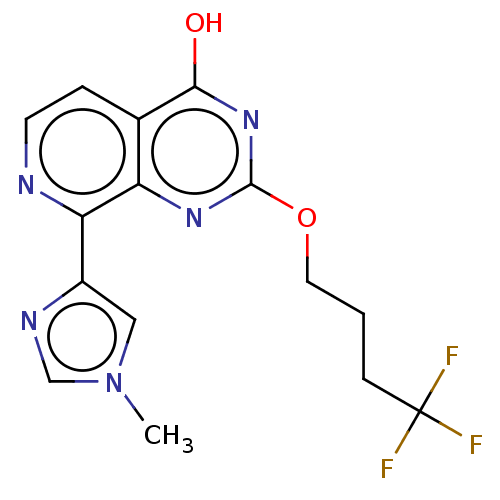
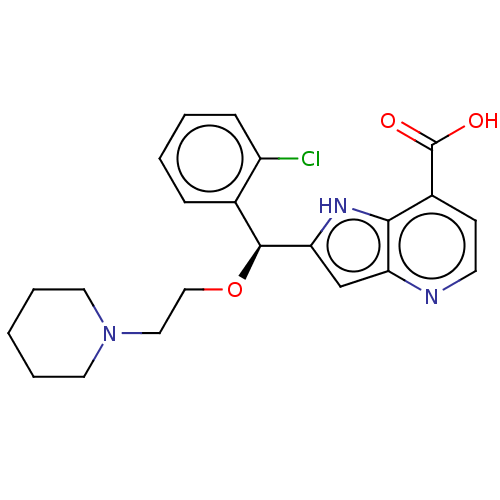
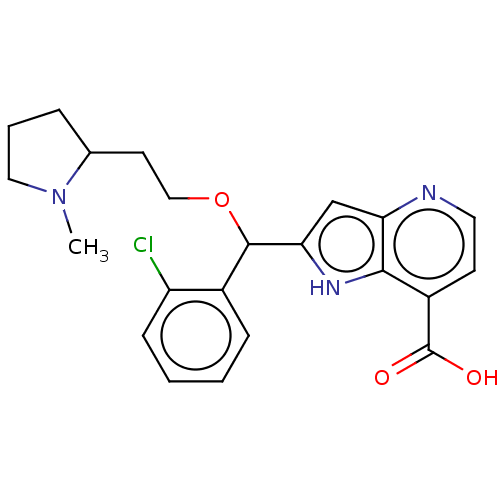
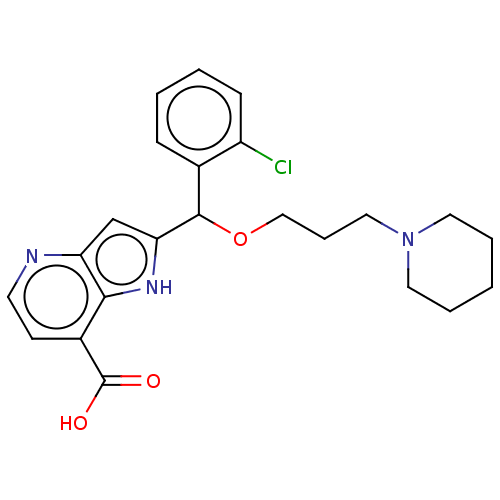
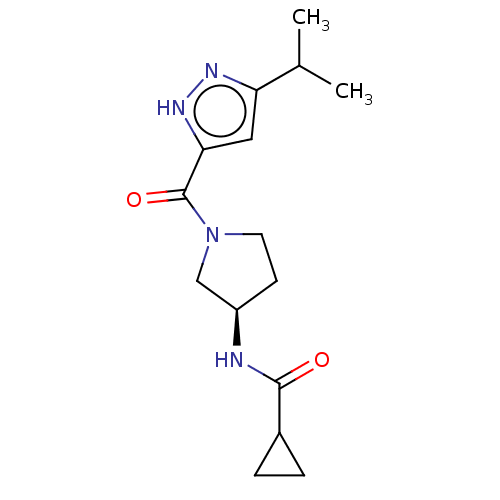
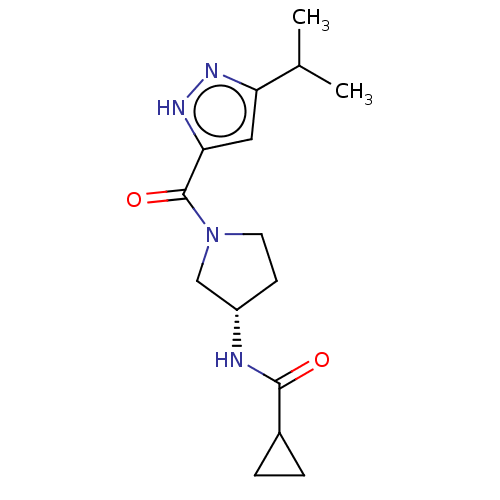
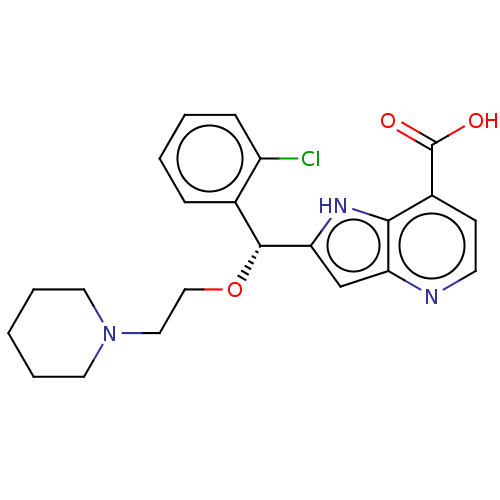
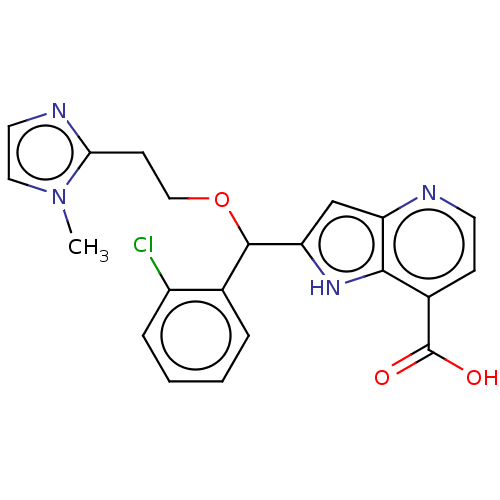
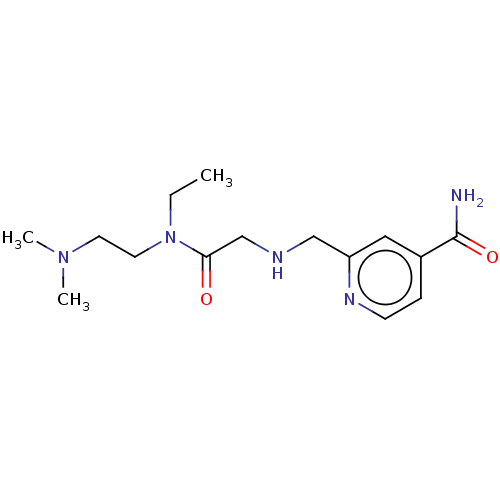
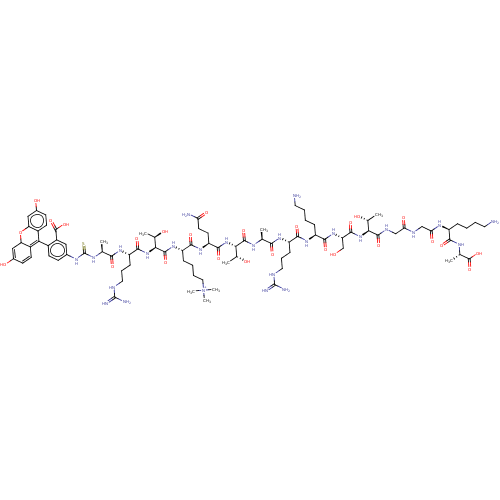
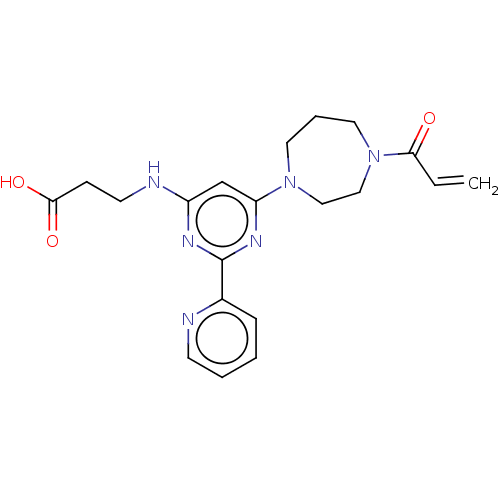
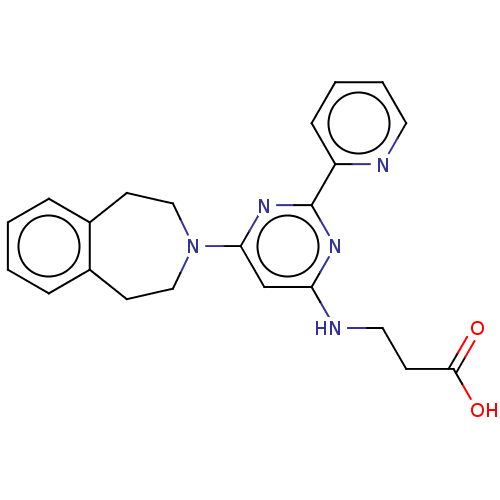
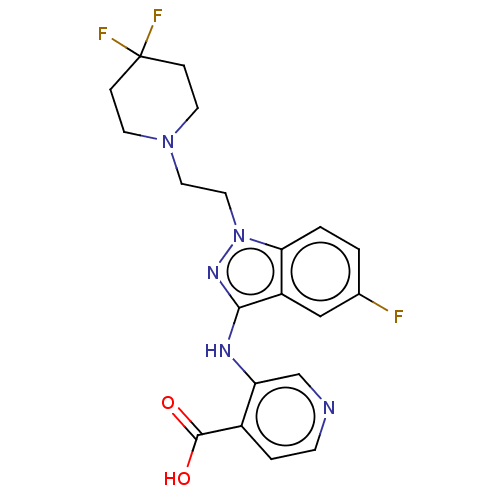
Affinity DataKd: 27nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
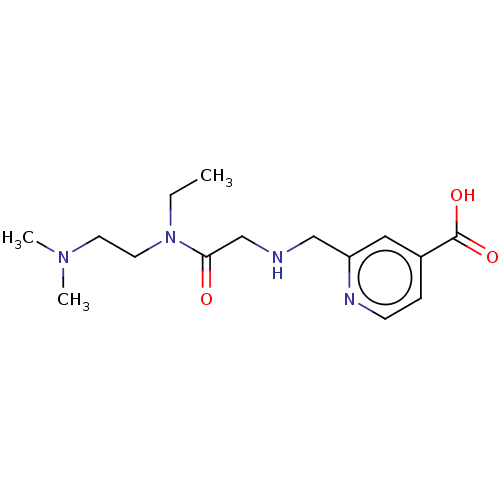
Affinity DataKd: 260nMAssay Description:Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assayMore data for this Ligand-Target Pair
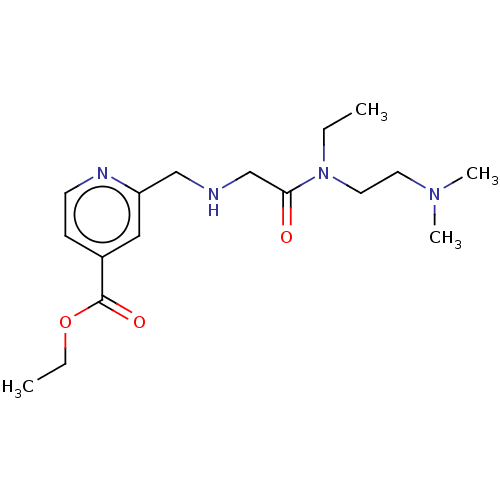
Affinity DataKd: 52nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair
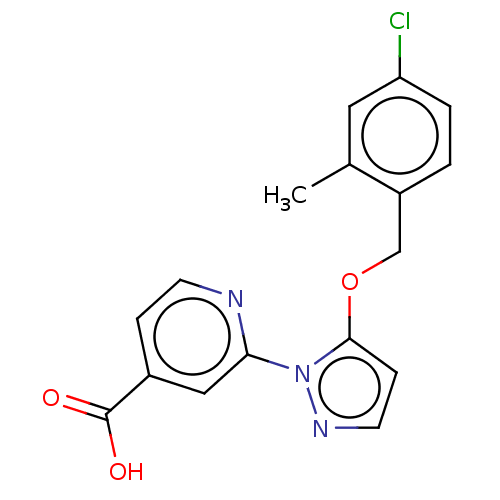
Affinity DataKd: >1.00E+4nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair
Affinity DataKd: 60nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair
Affinity DataKd: 360nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair
Affinity DataKd: 450nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair
Affinity DataKd: 1.20E+3nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair
Affinity DataKd: 7.00E+3nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair
Affinity DataKd: >4.00E+4nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 60nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 470nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 150nMAssay Description:Binding affinity at [3H]QNB and GppNHp radiolabeled muscarinic M2 receptor in rat heart.More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 250nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 2.40E+3nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: >1.20E+4nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 530nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 220nMAssay Description:Binding affinity at [3H]QNB radiolabeled muscarinic M2 receptor in rat heart.More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 210nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 410nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 2.70E+3nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 180nMAssay Description:Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by...More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 800nMAssay Description:Inhibition of JARID1A (unknown origin)More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 800nMAssay Description:Inhibition of JARID1A (unknown origin)More data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 9.00E+3nMAssay Description:Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assayMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 150nMAssay Description:Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assayMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 6.00E+3nMAssay Description:Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assayMore data for this Ligand-Target Pair
TargetLysine-specific demethylase 5A(Homo sapiens (Human))
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
Affinity DataKd: 1.20E+4nMAssay Description:Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assayMore data for this Ligand-Target Pair
Affinity DataKd: 46nMAssay Description:ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer...More data for this Ligand-Target Pair

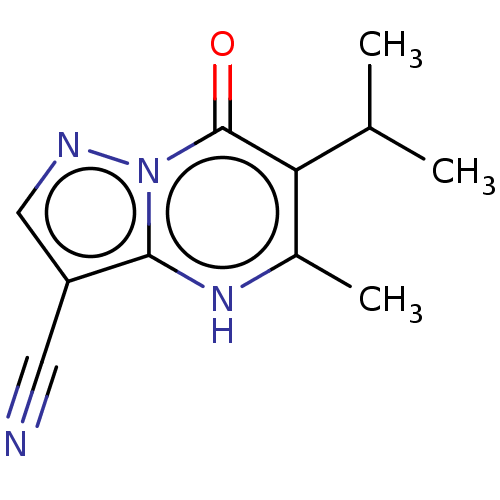
 3D Structure (crystal)
3D Structure (crystal)