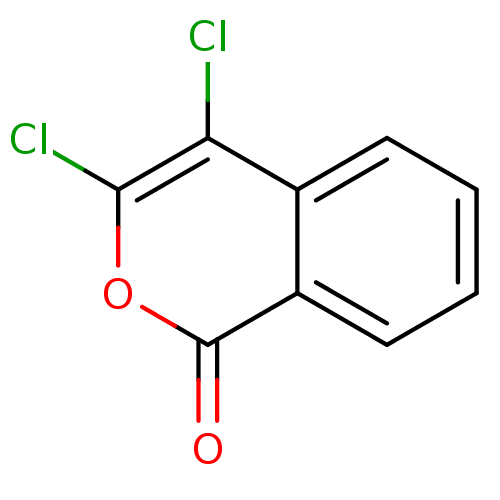
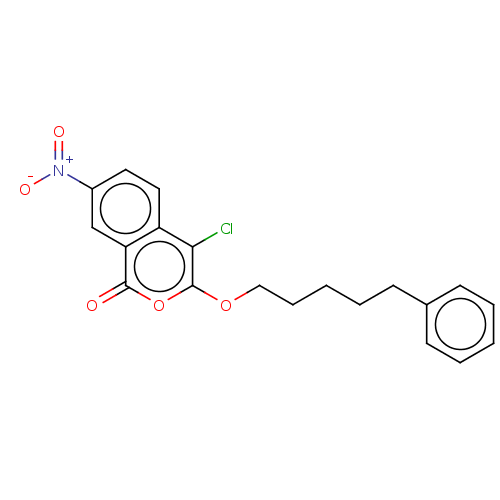
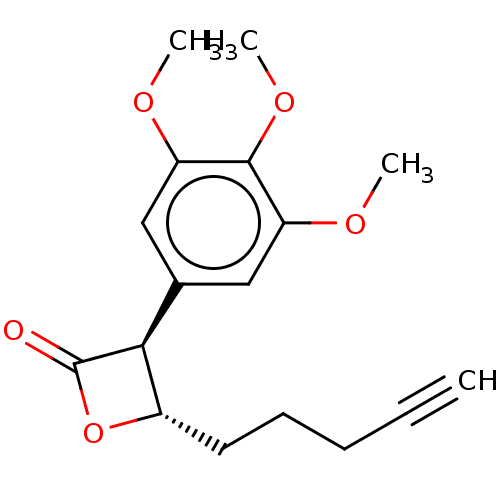
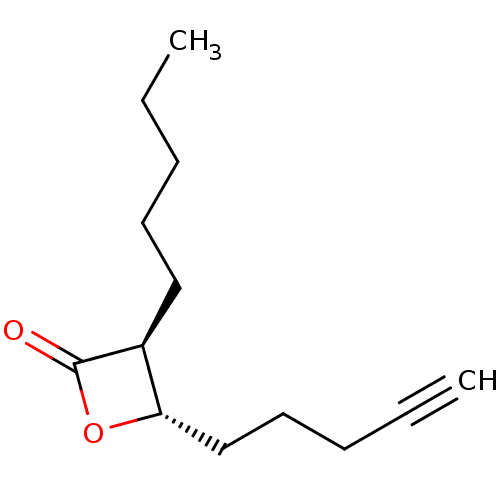
Found 25 hits of Enzyme Inhibition Constant Data
Found 25 hits of Enzyme Inhibition Constant Data
TargetRhomboid protease GlpG(Haemophilus influenzae (g-Proteobacteria))
Technische Universität München
Technische Universität München
Affinity DataIC50: 660nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid domain-containing protein(Thermotoga maritima (Thermotogales))
Technische Universität München
Technische Universität München
Affinity DataIC50: 1.00E+3nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid domain-containing protein(Aquifex aeolicus (Aquificales))
Technische Universität München
Technische Universität München
Affinity DataIC50: 1.10E+3nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid domain-containing protein(Aquifex aeolicus (Aquificales))
Technische Universität München
Technische Universität München
Affinity DataIC50: 1.70E+3nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid protease GluP [F193S,L370F,L379S,R499W](Bacillus subtilis (Firmicutes))
Technische Universität München
Technische Universität München
Affinity DataIC50: 4.10E+3nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid protease GlpG(Haemophilus influenzae (g-Proteobacteria))
Technische Universität München
Technische Universität München
Affinity DataIC50: 4.60E+3nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetAN1-type domain-containing protein(Archaeoglobus fulgidus (AfROM))
Technische Universität München
Technische Universität München
Affinity DataIC50: 5.00E+3nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetAN1-type domain-containing protein(Archaeoglobus fulgidus (AfROM))
Technische Universität München
Technische Universität München
Affinity DataIC50: 8.10E+3nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid domain-containing protein(Pyrococcus horikoshii (Euryarchaeotes))
Technische Universität München
Technische Universität München
Affinity DataIC50: 1.60E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 1.90E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 2.40E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid domain-containing protein(Pyrococcus horikoshii (Euryarchaeotes))
Technische Universität München
Technische Universität München
Affinity DataIC50: 2.50E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid protease GluP [F193S,L370F,L379S,R499W](Bacillus subtilis (Firmicutes))
Technische Universität München
Technische Universität München
Affinity DataIC50: 2.50E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid domain-containing protein(Thermotoga maritima (Thermotogales))
Technische Universität München
Technische Universität München
Affinity DataIC50: 2.70E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 2.70E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 2.80E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid domain-containing protein(Thermotoga maritima (Thermotogales))
Technische Universität München
Technische Universität München
Affinity DataIC50: 2.80E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 3.90E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 4.40E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid domain-containing protein(Thermotoga maritima (Thermotogales))
Technische Universität München
Technische Universität München
Affinity DataIC50: 4.80E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
TargetRhomboid protease GlpG(Haemophilus influenzae (g-Proteobacteria))
Technische Universität München
Technische Universität München
Affinity DataIC50: 6.50E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 7.00E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair
Affinity DataIC50: 7.60E+4nMpH: 7.4 T: 2°CAssay Description:We selected several inhibitor structures and determined their apparent IC50 by competitive ABPP.More data for this Ligand-Target Pair