Reaction Details  Report a problem with these data
Report a problem with these data
 Report a problem with these data
Report a problem with these dataTarget
Glycogen phosphorylase, muscle form
Ligand
BDBM50263769
Substrate
n/a
Meas. Tech.
ChEMBL_535893 (CHEMBL983534)
Ki
5100±n/a nM
Citation
 Somsák, L; Nagy, V; Vidal, S; Czifrák, K; Berzsényi, E; Praly, JP Novel design principle validated: glucopyranosylidene-spiro-oxathiazole as new nanomolar inhibitor of glycogen phosphorylase, potential antidiabetic agent. Bioorg Med Chem Lett 18:5680-3 (2008) [PubMed] Article
Somsák, L; Nagy, V; Vidal, S; Czifrák, K; Berzsényi, E; Praly, JP Novel design principle validated: glucopyranosylidene-spiro-oxathiazole as new nanomolar inhibitor of glycogen phosphorylase, potential antidiabetic agent. Bioorg Med Chem Lett 18:5680-3 (2008) [PubMed] ArticleMore Info.:
Target
Name:
Glycogen phosphorylase, muscle form
Synonyms:
Glycogen Phosphorylase (PYGM) | Glycogen phosphorylase a (RMGPa) | Glycogen phosphorylase, muscle form | Myophosphorylase | PYGM | PYGM_RABIT
Type:
Enzyme
Mol. Mass.:
97296.32
Organism:
Oryctolagus cuniculus (rabbit)
Description:
Phosphorylation of Ser-15 converts phosphorylase B (unphosphorylated) to phosphorylase A.
Residue:
843
Sequence:
MSRPLSDQEKRKQISVRGLAGVENVTELKKNFNRHLHFTLVKDRNVATPRDYYFALAHTVRDHLVGRWIRTQQHYYEKDPKRIYYLSLEFYMGRTLQNTMVNLALENACDEATYQLGLDMEELEEIEEDAGLGNGGLGRLAACFLDSMATLGLAAYGYGIRYEFGIFNQKICGGWQMEEADDWLRYGNPWEKARPEFTLPVHFYGRVEHTSQGAKWVDTQVVLAMPYDTPVPGYRNNVVNTMRLWSAKAPNDFNLKDFNVGGYIQAVLDRNLAENISRVLYPNDNFFEGKELRLKQEYFVVAATLQDIIRRFKSSKFGCRDPVRTNFDAFPDKVAIQLNDTHPSLAIPELMRVLVDLERLDWDKAWEVTVKTCAYTNHTVLPEALERWPVHLLETLLPRHLQIIYEINQRFLNRVAAAFPGDVDRLRRMSLVEEGAVKRINMAHLCIAGSHAVNGVARIHSEILKKTIFKDFYELEPHKFQNKTNGITPRRWLVLCNPGLAEIIAERIGEEYISDLDQLRKLLSYVDDEAFIRDVAKVKQENKLKFAAYLEREYKVHINPNSLFDVQVKRIHEYKRQLLNCLHVITLYNRIKKEPNKFVVPRTVMIGGKAAPGYHMAKMIIKLITAIGDVVNHDPVVGDRLRVIFLENYRVSLAEKVIPAADLSEQISTAGTEASGTGNMKFMLNGALTIGTMDGANVEMAEEAGEENFFIFGMRVEDVDRLDQRGYNAQEYYDRIPELRQIIEQLSSGFFSPKQPDLFKDIVNMLMHHDRFKVFADYEEYVKCQERVSALYKNPREWTRMVIRNIATSGKFSSDRTIAQYAREIWGVEPSRQRLPAPDEKIP
Inhibitor
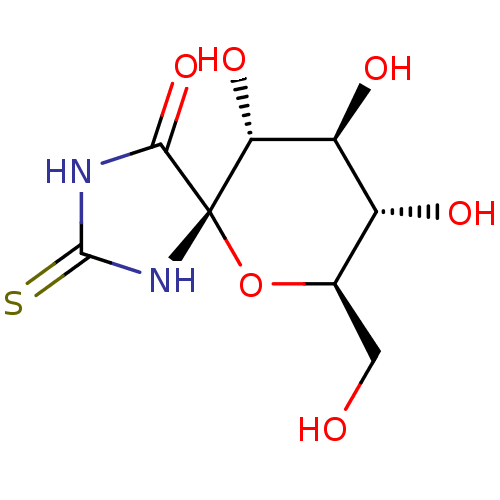
Name:
BDBM50263769
Synonyms:
(5S,7R,8S,9S,10R)-8,9,10-Trihydroxy-7-hydroxymethyl-2-thioxo-6-oxa-1,3-diaza-spiro[4.5]decan-4-one | (5S,7R,8S,9S,10R)-8,9,10-trihydroxy-7-(hydroxymethyl)-2-thioxo-6-oxa-1,3-diazaspiro[4.5]decan-4-one | 8,9,10-TRIHYDROXY-7-HYDROXYMETHYL-2-THIOXO-6-OXA-1,3-DIAZA-SPIRO[4.5]DECAN-4-ONE | CHEMBL489798
Type:
Small organic molecule
Emp. Form.:
C8H12N2O6S
Mol. Mass.:
264.256
SMILES:
OC[C@H]1O[C@@]2(NC(=S)NC2=O)[C@H](O)[C@@H](O)[C@@H]1O |r|