

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM22585 1H,3H,4H,7H-2,1,3,5,7-imidazo[4,5-c][1,2,6]thiadiazine-2,2,4-trione::Heterocycle, 1
SMILES: O=C1NS(=O)(=O)Nc2nc[nH]c12
InChI Key: InChIKey=BSAXWMSAVVOOHO-UHFFFAOYSA-N
Data: 4 KI
PDB links: 2 PDB IDs match this monomer. 2 PDB IDs contain this monomer as substructures. 2 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP Cyclohydrolase (IMPCH) (Homo sapiens (Human)) | BDBM22585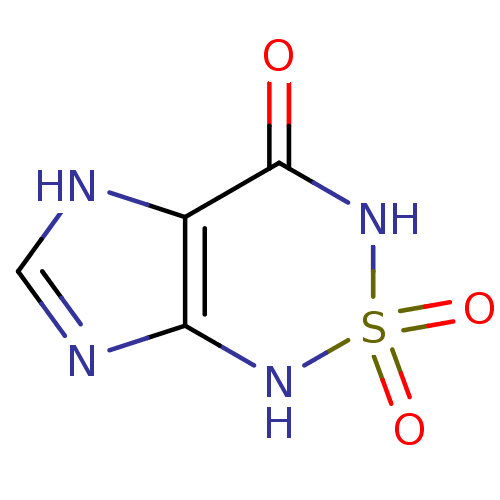 (1H,3H,4H,7H-2,1,3,5,7-imidazo[4,5-c][1,2,6]thiadia...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 130 | -9.29 | n/a | n/a | n/a | n/a | n/a | 7.4 | 22 |
The Scripps Research Institute | Assay Description The human ATIC enzyme was used for the inhibition assay using the spectrophotometric method monitoring the appearance of IMP by measuring absorbance ... | J Biol Chem 282: 13033-46 (2007) Article DOI: 10.1074/jbc.M607293200 BindingDB Entry DOI: 10.7270/Q2GM85MB | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glycinamide ribonucleotide formyltransferase (GARFTase) (Homo sapiens (Human)) | BDBM22585 (1H,3H,4H,7H-2,1,3,5,7-imidazo[4,5-c][1,2,6]thiadia...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | >1.00E+5 | >-5.47 | n/a | n/a | n/a | n/a | n/a | 7.5 | 26 |
The Scripps Research Institute | Assay Description Assays were initiated by the addition of GAR to the reaction mixture containing GAR Tfase, test compounds, and cofactor. The assay monitors the defor... | J Biol Chem 282: 13033-46 (2007) Article DOI: 10.1074/jbc.M607293200 BindingDB Entry DOI: 10.7270/Q2GM85MB | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| AICAR Tfase (Homo sapiens (Human)) | BDBM22585 (1H,3H,4H,7H-2,1,3,5,7-imidazo[4,5-c][1,2,6]thiadia...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | >1.00E+5 | >-5.40 | n/a | n/a | n/a | n/a | n/a | 7.5 | 22 |
The Scripps Research Institute | Assay Description Recombinant human AICAR Tfase was used in the inhibition assay. The reaction was monitored at 298 nm by measuring the increase in absorbance correspo... | J Biol Chem 282: 13033-46 (2007) Article DOI: 10.1074/jbc.M607293200 BindingDB Entry DOI: 10.7270/Q2GM85MB | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| GAR Tfase (Escherichia coli (strain K12)) | BDBM22585 (1H,3H,4H,7H-2,1,3,5,7-imidazo[4,5-c][1,2,6]thiadia...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | >1.00E+5 | >-5.47 | n/a | n/a | n/a | n/a | n/a | 7.5 | 26 |
The Scripps Research Institute | Assay Description Assays were initiated by the addition of GAR to the reaction mixture containing GAR Tfase, test compounds, and cofactor. The assay monitors the defor... | J Biol Chem 282: 13033-46 (2007) Article DOI: 10.1074/jbc.M607293200 BindingDB Entry DOI: 10.7270/Q2GM85MB | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||