

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM50200120 CHEMBL260091::CHIR-090::US10875832, Compound ChIR-090
SMILES: C[C@@H](O)[C@H](NC(=O)c1ccc(cc1)C#Cc1ccc(CN2CCOCC2)cc1)C(=O)NO
InChI Key: InChIKey=FQYBTYFKOHPWQT-VGSWGCGISA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UDP-3-O-acyl-GlcNAc deacetylase (Escherichia coli) | BDBM50200120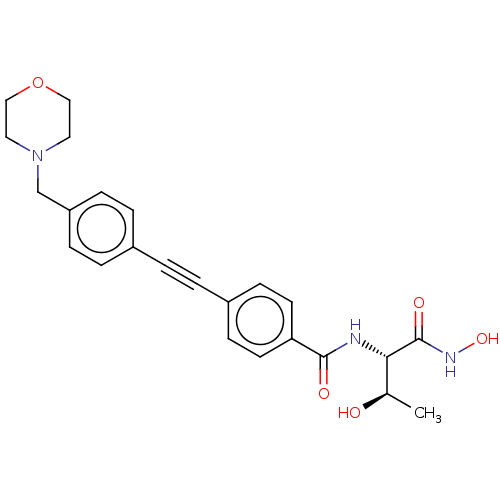 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | 0.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Duke University Medical Center Curated by ChEMBL | Assay Description Inhibition of Escherichia coli LpxC | Proc Natl Acad Sci U S A 104: 18433-8 (2007) Article DOI: 10.1073/pnas.0709412104 BindingDB Entry DOI: 10.7270/Q2SB48HR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (Escherichia coli) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | 0.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Duke University Curated by ChEMBL | Assay Description Inhibition of Escherichia coli LpxC enzyme | J Med Chem 56: 6954-6966 (2013) Article DOI: 10.1021/jm4007774 BindingDB Entry DOI: 10.7270/Q28K7D11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (Escherichia coli) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | 2 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Inhibition of Escherichia coli LpxC | Bioorg Med Chem Lett 22: 2536-43 (2012) Article DOI: 10.1016/j.bmcl.2012.01.140 BindingDB Entry DOI: 10.7270/Q2JS9T9W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (Escherichia coli) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | 2 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Escherichia coli LpxC | Bioorg Med Chem Lett 21: 1155-61 (2011) Article DOI: 10.1016/j.bmcl.2010.12.111 BindingDB Entry DOI: 10.7270/Q2Q52SFZ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-N-acetylglucosamine deacetylase (Rhizobium leguminosarum bv. trifolii (strain WSM13...) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | Article PubMed | 3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Duke University Medical Center Curated by ChEMBL | Assay Description Inhibition of Rhizobium leguminosarum LpxC W206Q/S214G mutant | Proc Natl Acad Sci U S A 104: 18433-8 (2007) Article DOI: 10.1073/pnas.0709412104 BindingDB Entry DOI: 10.7270/Q2SB48HR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| UDP-3-O-acyl-N-acetylglucosamine deacetylase (Rhizobium leguminosarum bv. trifolii (strain WSM13...) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | Article PubMed | 3.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Duke University Medical Center Curated by ChEMBL | Assay Description Inhibition of Rhizobium leguminosarum LpxC S214G mutant | Proc Natl Acad Sci U S A 104: 18433-8 (2007) Article DOI: 10.1073/pnas.0709412104 BindingDB Entry DOI: 10.7270/Q2SB48HR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (Escherichia coli) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | 5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Duke University Medical Center Curated by ChEMBL | Assay Description Inhibition of Escherichia coli LpxC Q202W/G210S mutant | Proc Natl Acad Sci U S A 104: 18433-8 (2007) Article DOI: 10.1073/pnas.0709412104 BindingDB Entry DOI: 10.7270/Q2SB48HR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (Escherichia coli) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | 8 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Institut f£r Pharmazeutische und Medizinische Chemie der Westf£lischen Wilhelms-Universit£t M£nster Curated by ChEMBL | Assay Description Inhibition of Escherichia coli LpxC using UDP-3-O-[(R)-3-hydroxymyristoyl]-N-acetylglucosamine as substrate incubated for 30 mins prior to enzyme add... | Bioorg Med Chem 22: 1016-28 (2014) Article DOI: 10.1016/j.bmc.2013.12.057 BindingDB Entry DOI: 10.7270/Q2X92F7C | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (LpxC) (Pseudomonas aeruginosa) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 0.480 | n/a | n/a | n/a | n/a | n/a |
Novartis Institutes for Biomedical Research Curated by ChEMBL | Assay Description Binding affinity to Pseudomonas aeruginosa PAO1 NB52019 LpxC after 60 to 420 secs by SPR method | J Med Chem 61: 9360-9370 (2018) Article DOI: 10.1021/acs.jmedchem.8b01287 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (Escherichia coli) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | 58 | n/a | n/a | n/a | n/a | n/a | n/a |
Institut f£r Pharmazeutische und Medizinische Chemie der Westf£lischen Wilhelms-Universit£t M£nster Curated by ChEMBL | Assay Description Inhibition of Escherichia coli LpxC using UDP-3-O-[(R)-3-hydroxymyristoyl]-N-acetylglucosamine as substrate incubated for 30 mins prior to enzyme add... | Bioorg Med Chem 22: 1016-28 (2014) Article DOI: 10.1016/j.bmc.2013.12.057 BindingDB Entry DOI: 10.7270/Q2X92F7C | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (Escherichia coli) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | 3 | n/a | n/a | n/a | n/a | n/a | n/a |
Kyorin Pharmaceutical Co., Ltd. Curated by ChEMBL | Assay Description Inhibition of Escherichia coli LpxC using UDP-3-O-(R-3-hydroxymyristoyl)GlcNAc as substrate after 60 mins by OPA reagent based fluorescence assay | Bioorg Med Chem Lett 27: 1045-1049 (2017) Article DOI: 10.1016/j.bmcl.2016.12.059 BindingDB Entry DOI: 10.7270/Q2MC9313 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (Escherichia coli) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | 3 | n/a | n/a | n/a | n/a | n/a | n/a |
Kyorin Pharmaceutical Co., Ltd. Curated by ChEMBL | Assay Description Inhibition of Escherichia coli LpxC using UDP-3-O-(R-3-hydroxymyristoyl)GlcNAc as substrate measured after 60 mins by fluorescence analysis | ACS Med Chem Lett 7: 623-8 (2016) Article DOI: 10.1021/acsmedchemlett.6b00057 BindingDB Entry DOI: 10.7270/Q2DV1NWV | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-N-acetylglucosamine deacetylase () | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | US Patent | n/a | n/a | <1 | n/a | n/a | n/a | n/a | n/a | n/a |
FORGE THERAPEUTICS, INC. US Patent | Assay Description LpxC inhibition assays were performed using liquid chromatography with tandem mass spectrometry. Assays were performed, in duplicate, in opaque, 96-w... | US Patent US10875832 (2020) | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (LpxC) (Pseudomonas aeruginosa) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | <2.10 | n/a | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Curated by ChEMBL | Assay Description Inhibition of wild-type Pseudomonas aeruginosa PAO1 LpxC assessed as deacetylation preincubated for 30 mins followed by addition of UDP-3-O-(R-hydrox... | Bioorg Med Chem 24: 6320-6331 (2016) Article DOI: 10.1016/j.bmc.2016.05.004 BindingDB Entry DOI: 10.7270/Q2VQ34NX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-3-O-acyl-GlcNAc deacetylase (LpxC) (Pseudomonas aeruginosa) | BDBM50200120 (CHEMBL260091 | CHIR-090 | US10875832, Compound ChI...) | PDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | 310 | n/a | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Curated by ChEMBL | Assay Description Inhibition of Pseudomonas aeruginosa LpxC using UDP-3-O-(R-3-hydroxydecanoyl)-N-acetylglucosamine as substrate preincubated for 20 mins before substr... | ACS Med Chem Lett 5: 1213-8 (2014) Article DOI: 10.1021/ml500210x BindingDB Entry DOI: 10.7270/Q2F76GKR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||