

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207976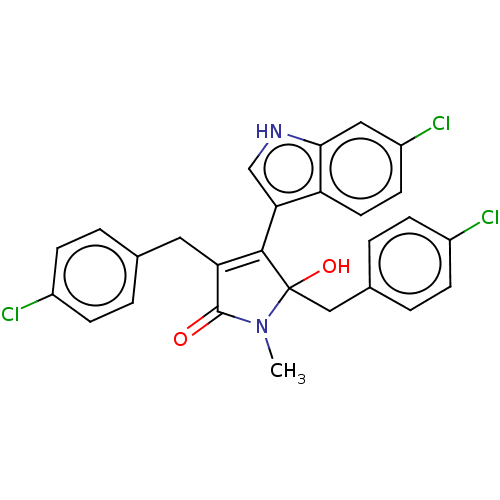 (N-Methyl-3,5-di(4-chlorobenzyl)-4-(6-chloro-1H-ind...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 500 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207978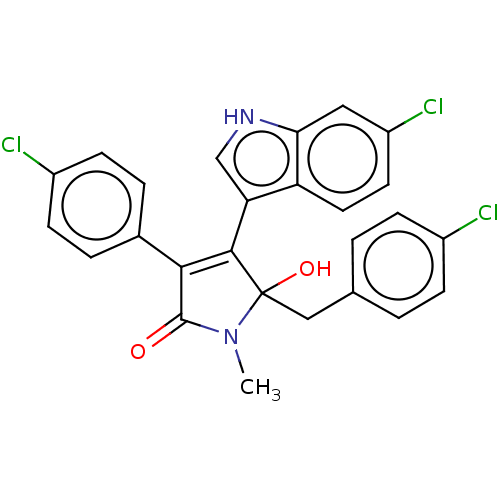 (N-Methyl-5-(4-chlorobenzyl)-4-(6-chloro-1H-indol-3...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.87E+4 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207979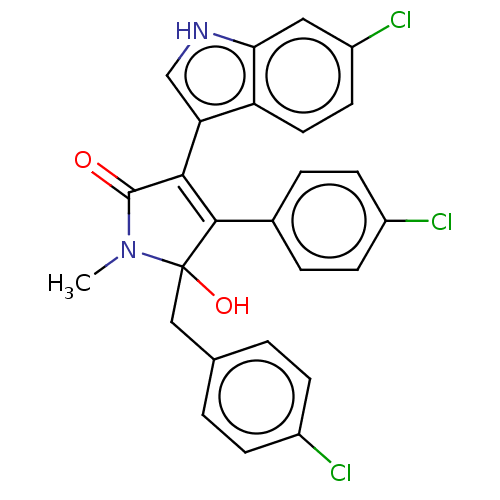 (N-methyl-5-(4-chlorobenzyl)-3-(6-chloro-1H-indol-3...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 2.77E+4 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207980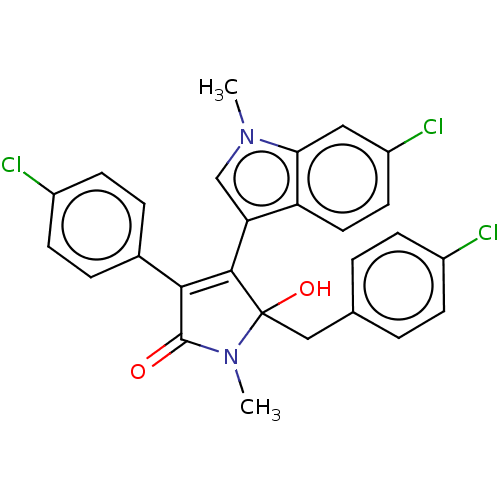 (N-Methyl-5-(4-chlorobenzyl)-4-(6-chloro-1-methyl-1...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 4.05E+4 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207981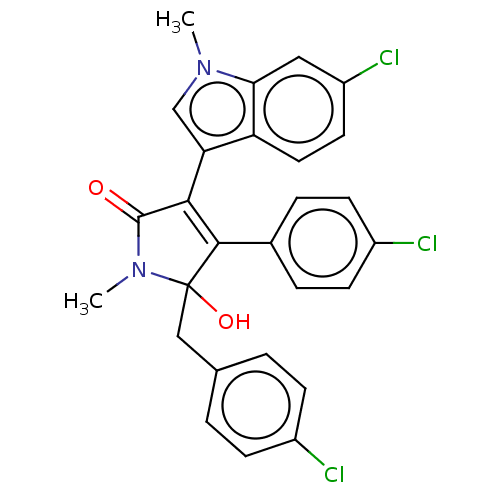 (N-methyl-5-(4-chlorobenzyl)-3-(6-chloro-1-methyl-1...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 3.04E+4 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207982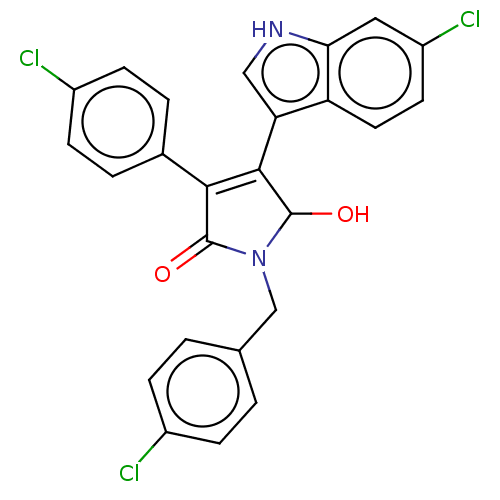 (N-(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)- 3-(...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 2.44E+4 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207988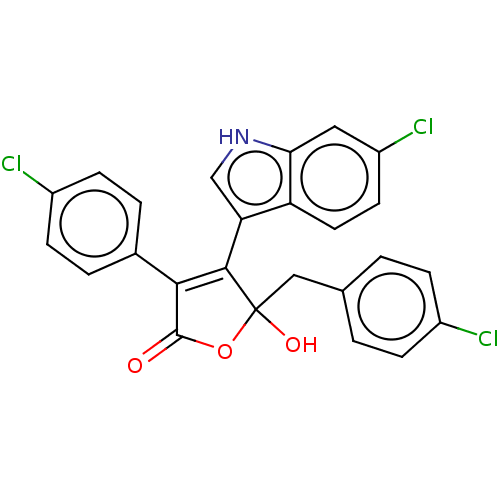 (5-(4-Chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-3-(4...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.30E+3 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207984 (3,5-Di(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 3.10E+3 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207985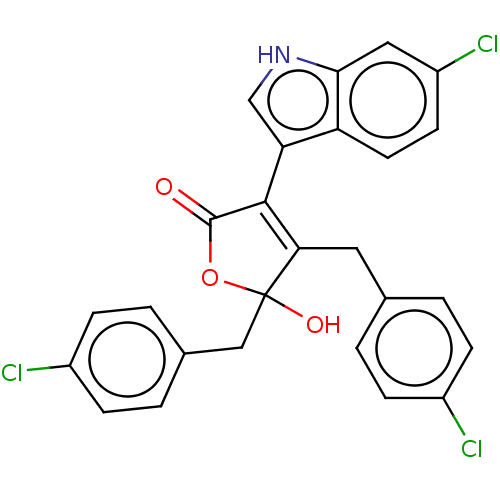 (4,5-di(4-chlorobenzyl)-3-(6-chloro-1H-indol-3-yl)-...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.50E+3 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207987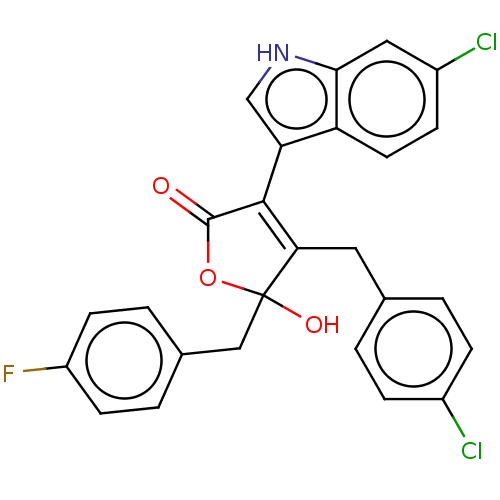 (4-(4-chlorobenzyl)-3-(6-chloro-1H-indol-3-yl)-5-(4...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 2.60E+4 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207989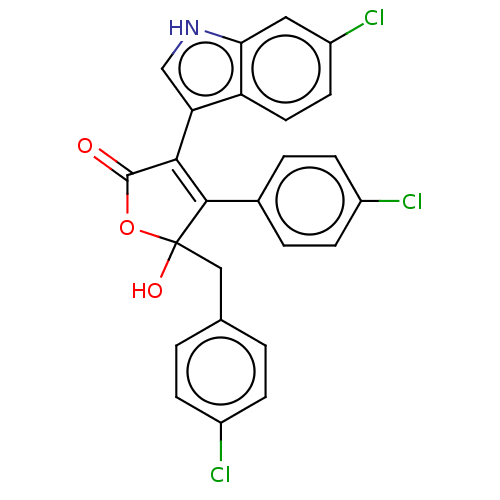 (5-(4-chlorobenzyl)-3-(6-chloro-1H-indol-3-yl)-4-(4...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 7.00E+3 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207986 (3-(4-Chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-(4...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 2.87E+4 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-118,T47W] (Homo sapiens (Human)) | BDBM207983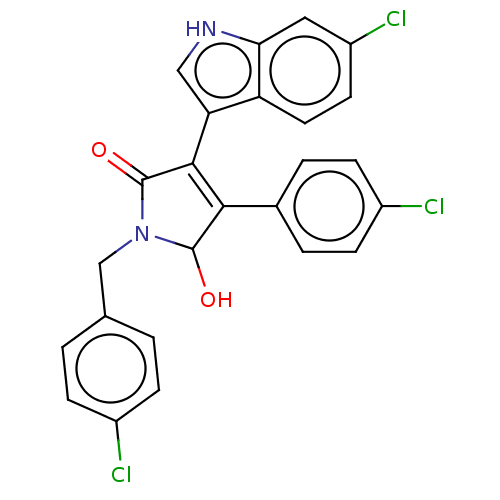 (N-(4-Chlorobenzyl)-3-(6-chloro-1H-indol-3-yl)-4-(4...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 2.10E+3 | n/a | n/a | n/a | 7.4 | 25 |
Jagiellonian University | Assay Description MST (NanoTemper Technologies GmbH) was used to determine the binding affinities between Mdm2 (residues 1−118 T47W; 500 nM) and inhibitors. T47W... | ACS Chem Biol 11: 3310-3318 (2016) Article DOI: 10.1021/acschembio.6b00596 BindingDB Entry DOI: 10.7270/Q2K35SH2 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||