 Found 15 hits of ki for monomerid = 50229787
Found 15 hits of ki for monomerid = 50229787 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787
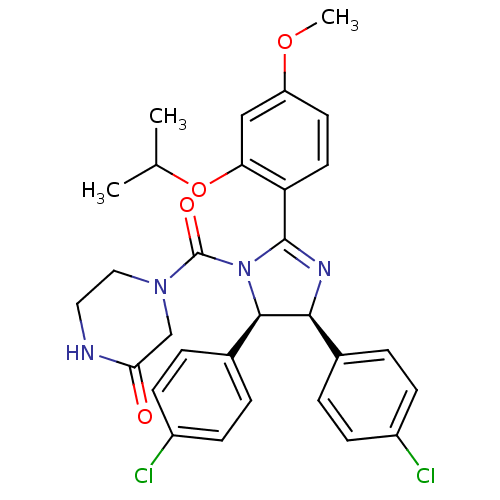
((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 6.40 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Novartis Institutes for BioMedical Research
Curated by ChEMBL
| Assay Description
Inhibition of human MDM2 by TR-FRET assay |
J Med Chem 58: 6348-58 (2015)
Article DOI: 10.1021/acs.jmedchem.5b00810
BindingDB Entry DOI: 10.7270/Q2HQ41QV |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Canis lupus familiaris) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 11 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Novartis Institutes for BioMedical Research
Curated by ChEMBL
| Assay Description
Inhibition of dog MDM2 by TR-FRET assay |
J Med Chem 58: 6348-58 (2015)
Article DOI: 10.1021/acs.jmedchem.5b00810
BindingDB Entry DOI: 10.7270/Q2HQ41QV |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Mus musculus) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 20 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Novartis Institutes for BioMedical Research
Curated by ChEMBL
| Assay Description
Inhibition of mouse MDM2 by TR-FRET assay |
J Med Chem 58: 6348-58 (2015)
Article DOI: 10.1021/acs.jmedchem.5b00810
BindingDB Entry DOI: 10.7270/Q2HQ41QV |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 39 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00095
BindingDB Entry DOI: 10.7270/Q2V69PP3 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 43 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Naples "Federico II"
Curated by ChEMBL
| Assay Description
Binding affinity to 6xHis-taged MDM2 (unknown origin) expressed in Escherichia coli Gold (DE3) assessed as inhibition constant |
ACS Med Chem Lett 11: 1047-1053 (2020)
Article DOI: 10.1021/acsmedchemlett.9b00620
BindingDB Entry DOI: 10.7270/Q20K2D3J |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 59 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Dalian University of Technology
Curated by ChEMBL
| Assay Description
Displacement of FAM-p53TAD peptide from N-terminal 8x His-tagged human MDM2 (25 to 108 residues) expressed in Escherichia coli BL21 (DE3) after 30 mi... |
J Med Chem 59: 3152-62 (2016)
Article DOI: 10.1021/acs.jmedchem.5b01913
BindingDB Entry DOI: 10.7270/Q2S75KBK |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 90 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Second Military Medical University
Curated by ChEMBL
| Assay Description
Binding affinity to MDM2 (unknown origin) assessed as inhibition of interaction with p53 after 1 hr by fluorescence polarization binding assay |
Bioorg Med Chem Lett 24: 2648-50 (2014)
Article DOI: 10.1016/j.bmcl.2014.04.063
BindingDB Entry DOI: 10.7270/Q2QV3P22 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 110 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Chinese Academy of Medical Science and Peking Union Medical College
Curated by ChEMBL
| Assay Description
Binding affinity to MDM2 (1 to 118) (unknown origin) after 30 mins by fluorescence polarization assay |
Eur J Med Chem 81: 277-88 (2014)
Article DOI: 10.1016/j.ejmech.2014.05.027
BindingDB Entry DOI: 10.7270/Q2736SGW |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 121 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Inhibition of MDM2 binding domain assessed as inhibition of p53 binding to MDM2 after 1 hr by fluorescence polarization assay |
Eur J Med Chem 56: 10-16 (2012)
Article DOI: 10.1016/j.ejmech.2012.08.003
BindingDB Entry DOI: 10.7270/Q2K938SR |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 140 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
East China University of Science and Technology
Curated by ChEMBL
| Assay Description
Inhibition of MDM2 (unknown origin) preincubated for 30 mins by fluorescence polarization assay |
J Med Chem 61: 7245-7260 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00664
BindingDB Entry DOI: 10.7270/Q2R213Z9 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
| 150 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| Assay Description
In vitro antagonism of the 5-HT-3 receptor determined by inhibition of 5-HT-induced depolarization of the isolated rat vagus nerve. |
Citation and Details
|
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Induced myeloid leukemia cell differentiation protein Mcl-1
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| Article
PubMed
| >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Dalian University of Technology
Curated by ChEMBL
| Assay Description
Displacement of FAM-Bid peptide from N-terminal 8x His-tagged MCl-1 (unknown origin) expressed in Escherichia coli BL21 (DE3) after 30 mins by fluore... |
J Med Chem 59: 3152-62 (2016)
Article DOI: 10.1021/acs.jmedchem.5b01913
BindingDB Entry DOI: 10.7270/Q2S75KBK |
More data for this
Ligand-Target Pair | |
Apoptosis regulator Bcl-2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
UniProtKB/SwissProt
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| Article
PubMed
| >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Dalian University of Technology
Curated by ChEMBL
| Assay Description
Displacement of FAM-Bid peptide from N-terminal 8x His-tagged Bcl-2 (unknown origin) expressed in Escherichia coli BL21 (DE3) after 30 mins by fluore... |
J Med Chem 59: 3152-62 (2016)
Article DOI: 10.1021/acs.jmedchem.5b01913
BindingDB Entry DOI: 10.7270/Q2S75KBK |
More data for this
Ligand-Target Pair | |
Protein Mdm4
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
| 6.90E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| Assay Description
Tested for ability to inhibit human erythrocyte carbonic-anhydrase-II- (HCA II) catalyzed carbon dioxide hydration |
Citation and Details
|
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50229787

((4S,5R)-Nutlin-3 | (rac)-(4,5-bis(4-chlorophenyl)-...)Show SMILES COc1ccc(C2=N[C@H]([C@H](N2C(=O)N2CCNC(=O)C2)c2ccc(Cl)cc2)c2ccc(Cl)cc2)c(OC(C)C)c1 |t:6| Show InChI InChI=1S/C30H30Cl2N4O4/c1-18(2)40-25-16-23(39-3)12-13-24(25)29-34-27(19-4-8-21(31)9-5-19)28(20-6-10-22(32)11-7-20)36(29)30(38)35-15-14-33-26(37)17-35/h4-13,16,18,27-28H,14-15,17H2,1-3H3,(H,33,37)/t27-,28+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
| 7.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| Assay Description
Tested for ability to inhibit human erythrocyte carbonic-anhydrase-II- (HCA II) catalyzed carbon dioxide hydration |
Citation and Details
|
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data