

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470392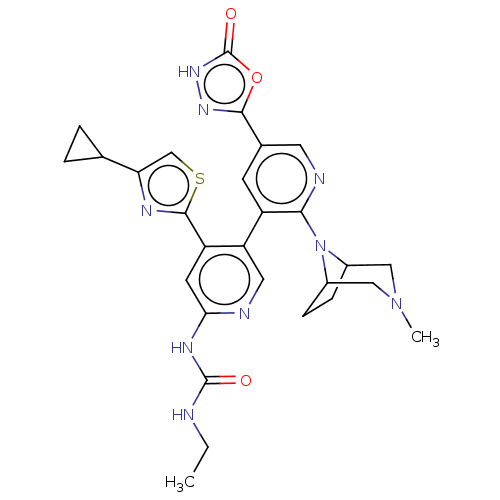 (CHEMBL4283208) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 2 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470399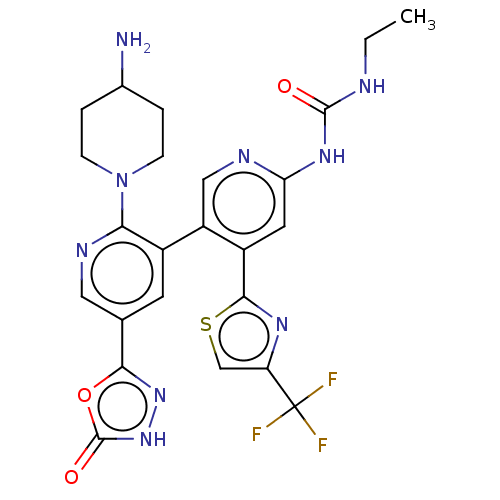 (CHEMBL4286573) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 2 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470406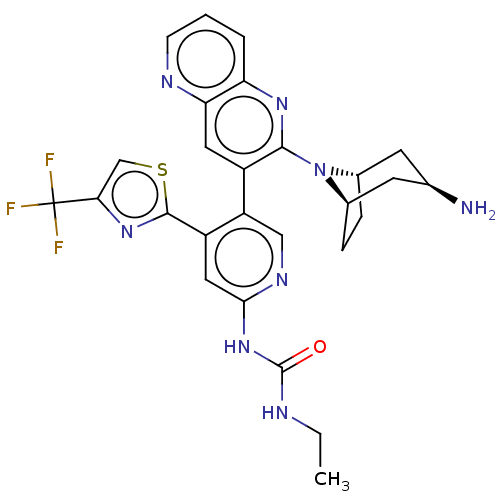 (CHEMBL4295081) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 3 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470397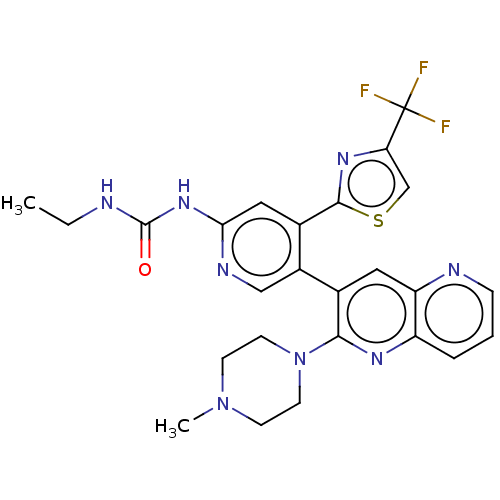 (CHEMBL4294536) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 3 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470393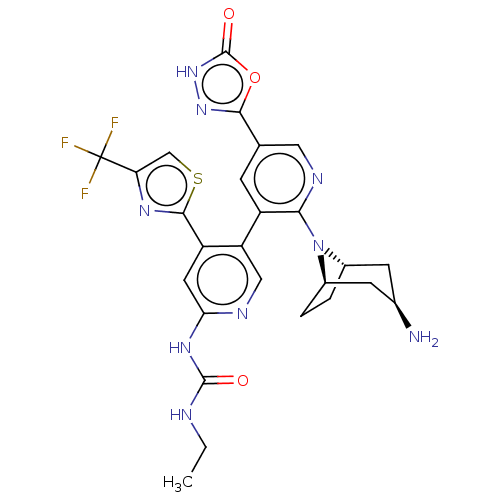 (CHEMBL4291061) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 4 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470407 (CHEMBL4286625) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 4 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470401 (CHEMBL4290062) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 5 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470396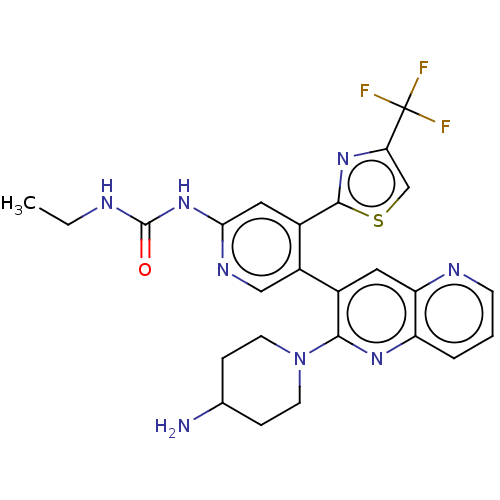 (CHEMBL4293447) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 6 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470403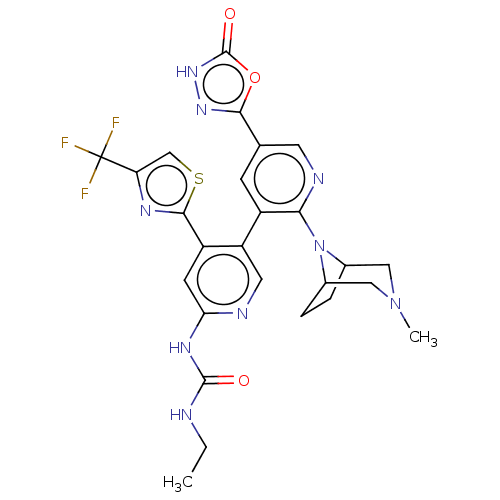 (CHEMBL4294467) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | 6 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470394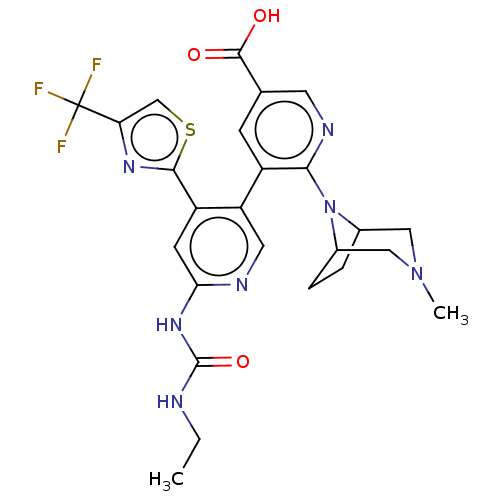 (CHEMBL4281775) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 6 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50006565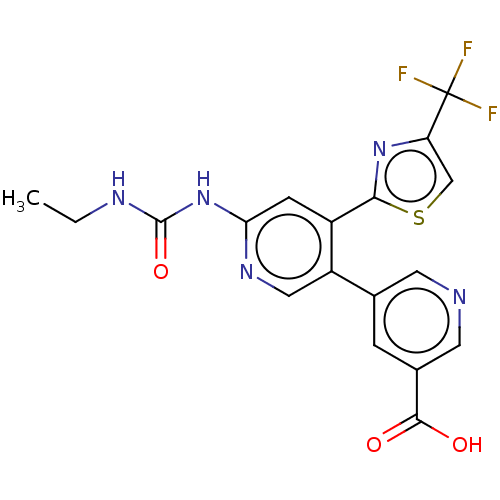 (CHEMBL3235085) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 6 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470404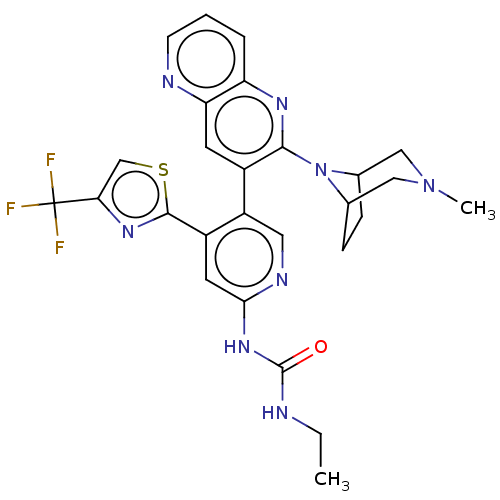 (CHEMBL4279382) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 7 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470402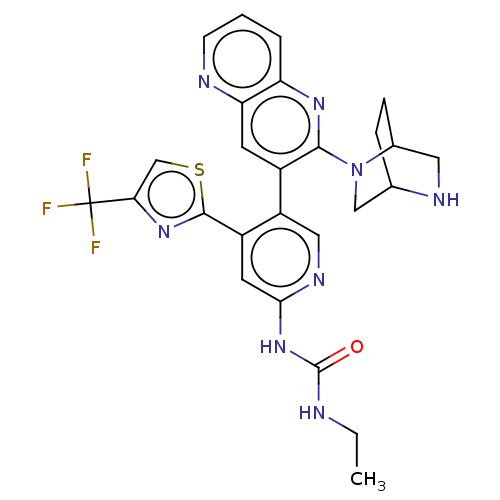 (CHEMBL4278317) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 8 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470395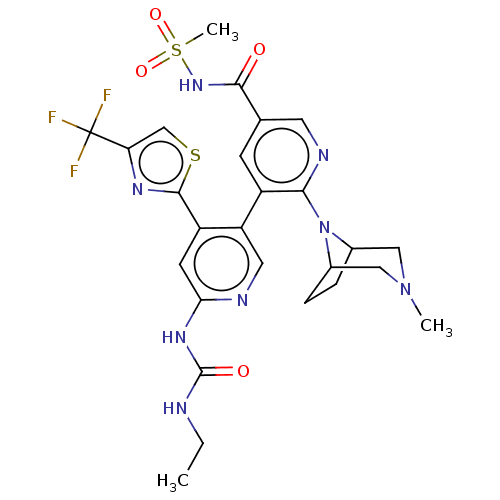 (CHEMBL4279313) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 8 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470405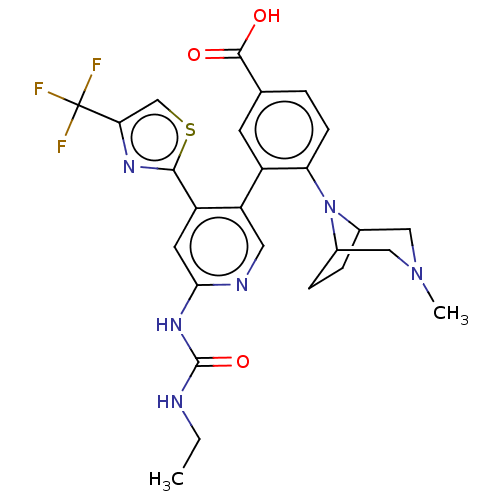 (CHEMBL4289650) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 9 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470400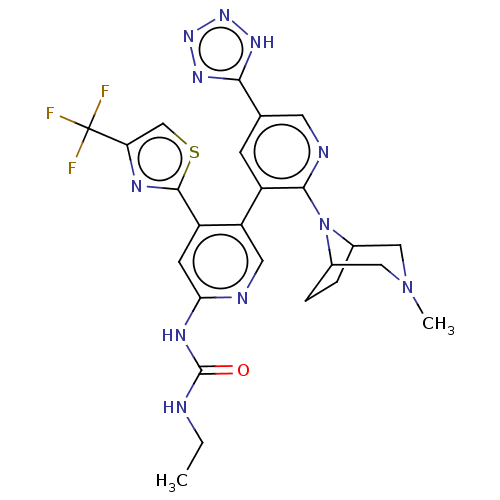 (CHEMBL4289998) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 9 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50408191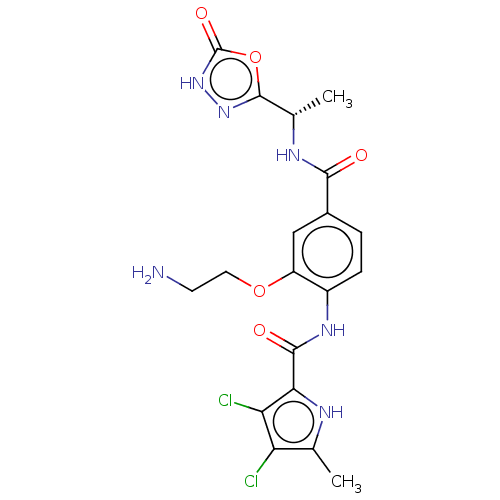 (CHEMBL4163243) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 13 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli C-terminal His6-tagged DNA gyrase B expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by ATP a... | Eur J Med Chem 154: 117-132 (2018) Article DOI: 10.1016/j.ejmech.2018.05.011 BindingDB Entry DOI: 10.7270/Q2CV4M9N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50470398 (CHEMBL4286232) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 14 | n/a | n/a | n/a | n/a | n/a | n/a |
Experimental Therapeutics Centre Curated by ChEMBL | Assay Description Inhibition of Escherichia coli GyrB (1 to 220 residues) expressed in Escherichia coli BL21(DE3) | Eur J Med Chem 157: 610-621 (2018) Article DOI: 10.1016/j.ejmech.2018.08.025 BindingDB Entry DOI: 10.7270/Q28P637N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330319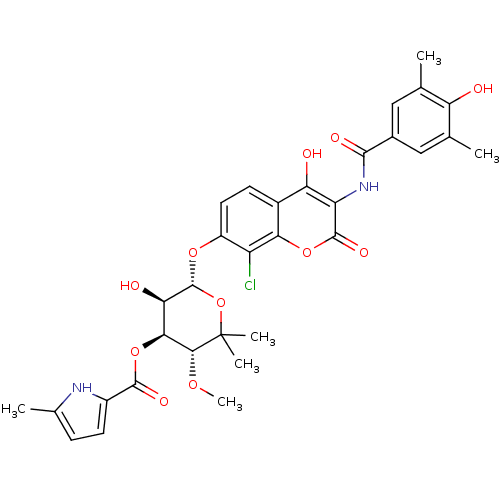 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 17 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330315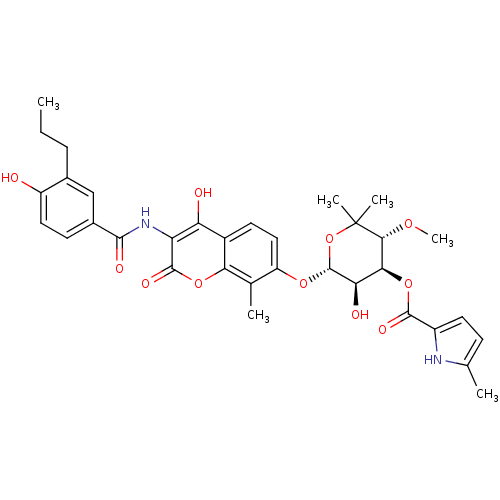 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 19 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330314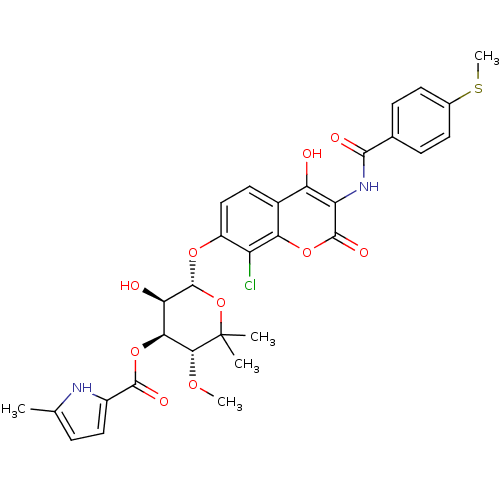 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-(methylth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 20 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330309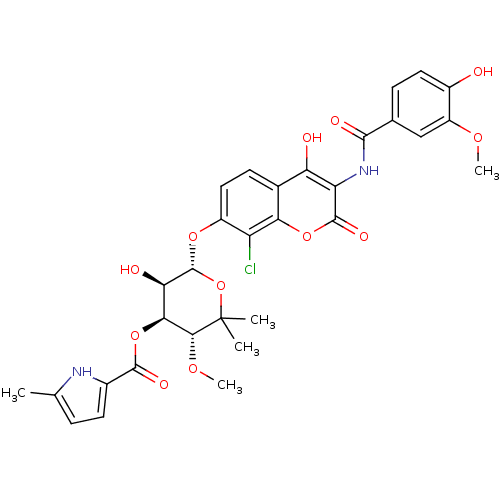 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 22 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330313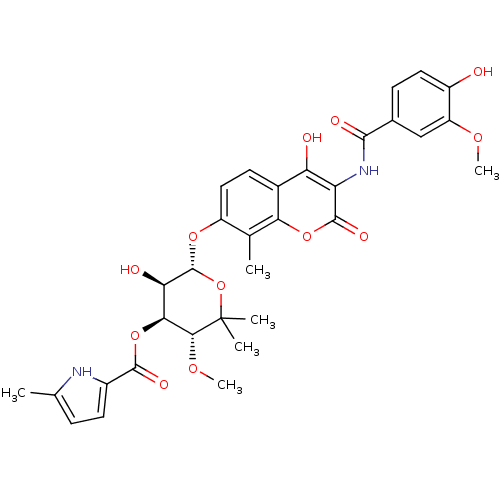 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 25 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50407015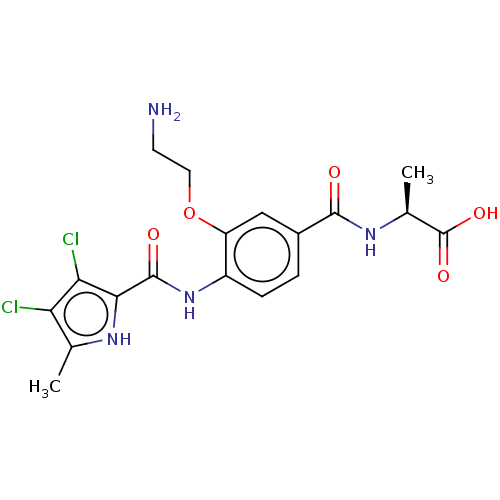 (CHEMBL4161729) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 28 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli C-terminal His6-tagged DNA gyrase B expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by ATP a... | Eur J Med Chem 154: 117-132 (2018) Article DOI: 10.1016/j.ejmech.2018.05.011 BindingDB Entry DOI: 10.7270/Q2CV4M9N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330307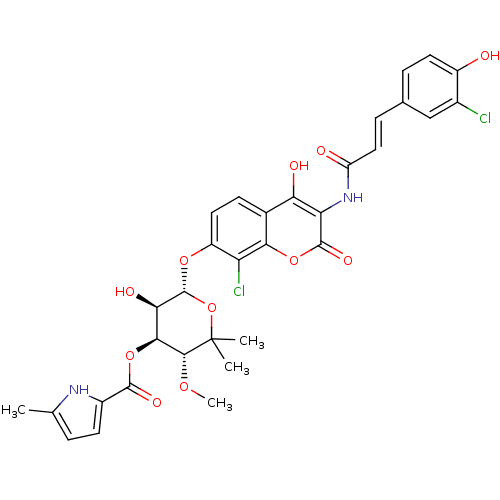 ((3R,4S,5R,6S)-6-(8-chloro-3-((E)-3-(3-chloro-4-hyd...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 32 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50198238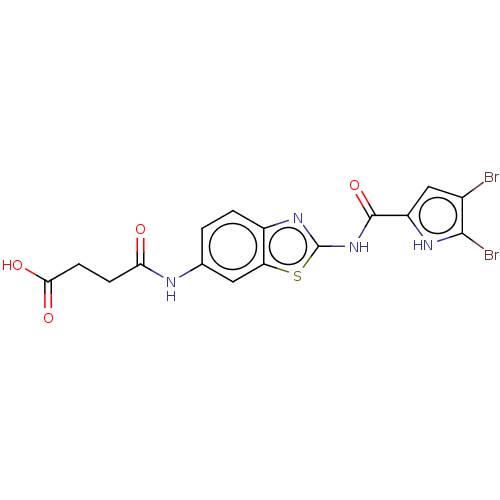 (CHEMBL3903891) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 33 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli DNA gyrase B ATP binding site assessed as reduction in supercoiling of relaxed pNO1 DNA after 30 mins in presence of b... | J Med Chem 59: 8941-8954 (2016) Article DOI: 10.1021/acs.jmedchem.6b00864 BindingDB Entry DOI: 10.7270/Q25B04GJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50408188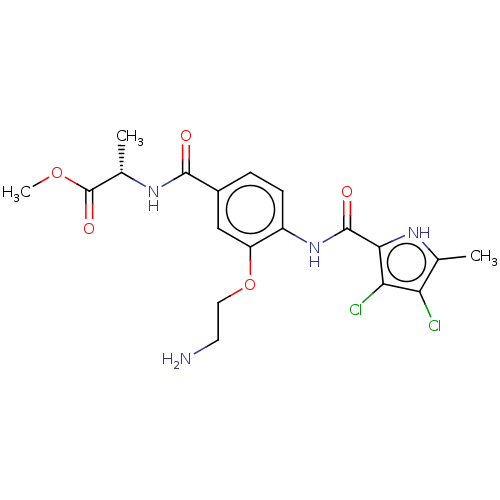 (CHEMBL4160665) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 34 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli C-terminal His6-tagged DNA gyrase B expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by ATP a... | Eur J Med Chem 154: 117-132 (2018) Article DOI: 10.1016/j.ejmech.2018.05.011 BindingDB Entry DOI: 10.7270/Q2CV4M9N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330310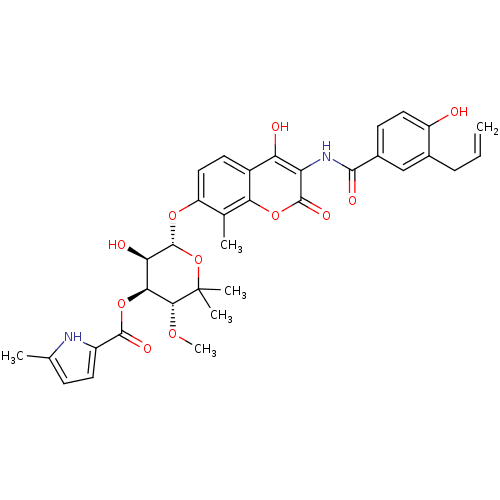 ((3R,4S,5R,6R)-6-(3-(3-allyl-4-hydroxybenzamido)-4-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 34 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330312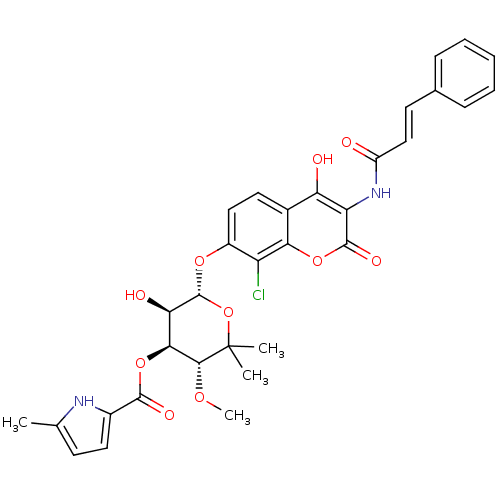 ((3R,4S,5R,6S)-6-(8-chloro-3-cinnamamido-4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 34 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50407009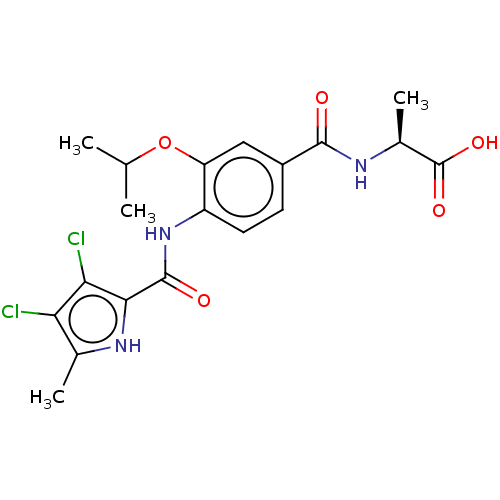 (CHEMBL4162886) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 38 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli C-terminal His6-tagged DNA gyrase B expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by ATP a... | Eur J Med Chem 154: 117-132 (2018) Article DOI: 10.1016/j.ejmech.2018.05.011 BindingDB Entry DOI: 10.7270/Q2CV4M9N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50198237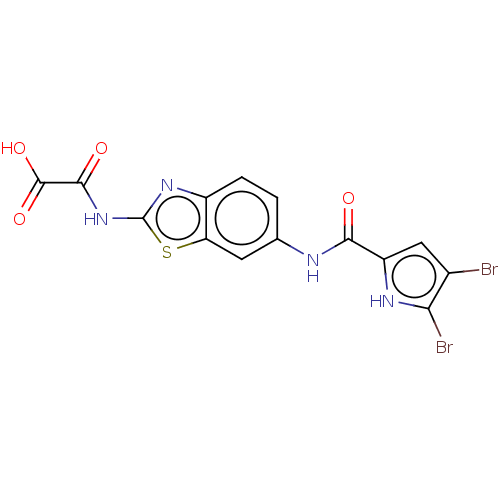 (CHEMBL3898662) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 38 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli DNA gyrase B ATP binding site assessed as reduction in supercoiling of relaxed pNO1 DNA after 30 mins in presence of b... | J Med Chem 59: 8941-8954 (2016) Article DOI: 10.1021/acs.jmedchem.6b00864 BindingDB Entry DOI: 10.7270/Q25B04GJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330316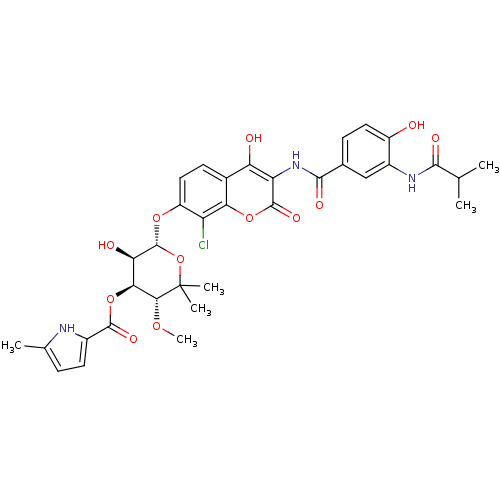 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 41 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50407460 (CHEMBL4175379) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 41 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli C-terminal His6-tagged DNA gyrase B expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by ATP a... | Eur J Med Chem 154: 117-132 (2018) Article DOI: 10.1016/j.ejmech.2018.05.011 BindingDB Entry DOI: 10.7270/Q2CV4M9N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330302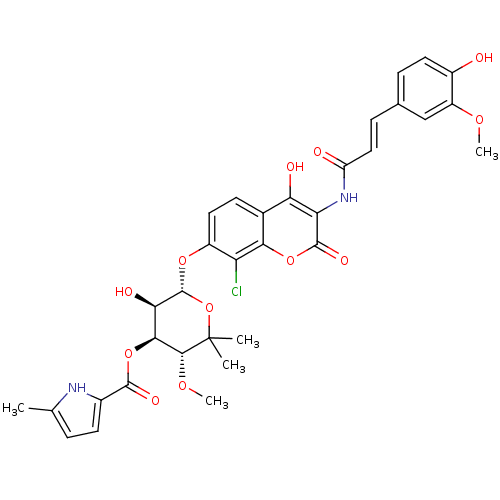 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-((E)-3-(4-hy...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 43 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330306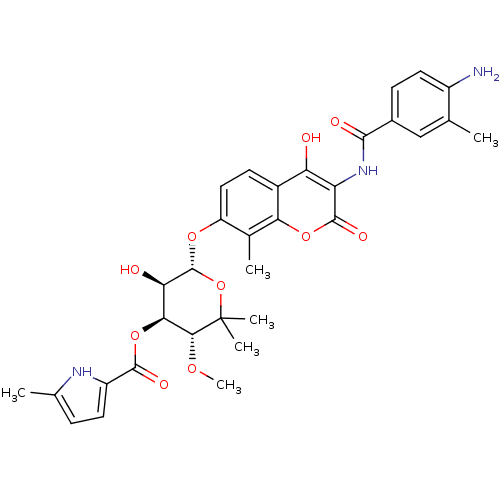 ((3R,4S,5R,6R)-6-(3-(4-amino-3-methylbenzamido)-4-h...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 44 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50408203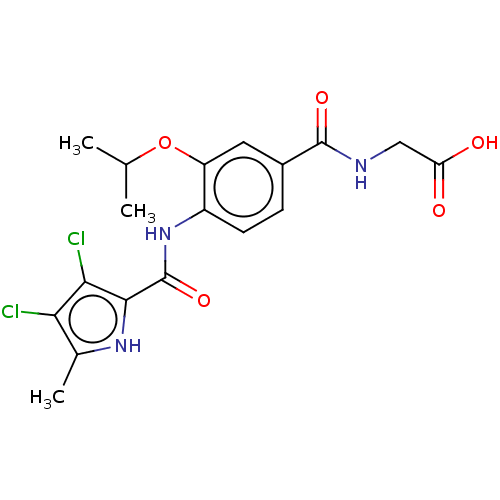 (CHEMBL4176270) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 47 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli C-terminal His6-tagged DNA gyrase B expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by ATP a... | Eur J Med Chem 154: 117-132 (2018) Article DOI: 10.1016/j.ejmech.2018.05.011 BindingDB Entry DOI: 10.7270/Q2CV4M9N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330304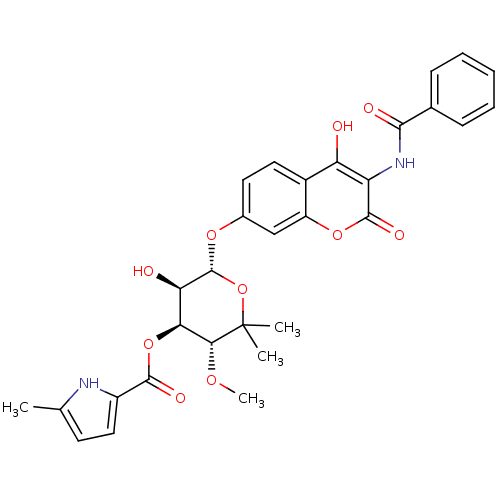 ((3R,4S,5R,6R)-6-(3-benzamido-4-hydroxy-2-oxo-2H-ch...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 50 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50198315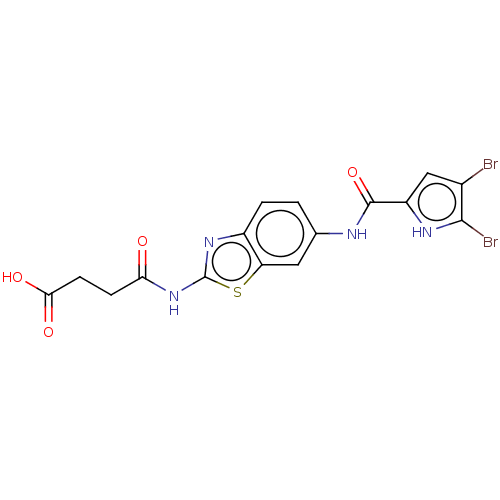 (CHEMBL3973476) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 57 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli DNA gyrase B ATP binding site assessed as reduction in supercoiling of relaxed pNO1 DNA after 30 mins in presence of b... | J Med Chem 59: 8941-8954 (2016) Article DOI: 10.1021/acs.jmedchem.6b00864 BindingDB Entry DOI: 10.7270/Q25B04GJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50198240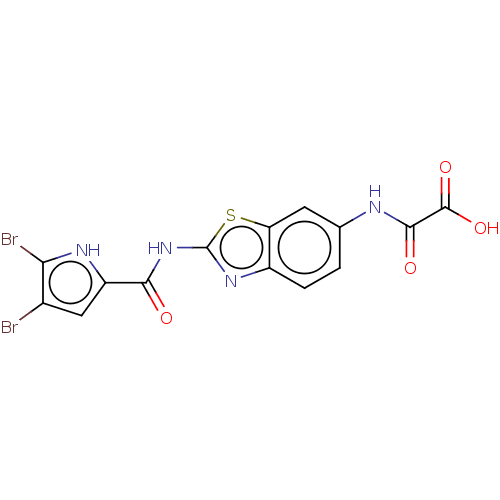 (CHEMBL3908487) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 58 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli DNA gyrase B ATP binding site assessed as reduction in supercoiling of relaxed pNO1 DNA after 30 mins in presence of b... | J Med Chem 59: 8941-8954 (2016) Article DOI: 10.1021/acs.jmedchem.6b00864 BindingDB Entry DOI: 10.7270/Q25B04GJ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330319 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 65 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330318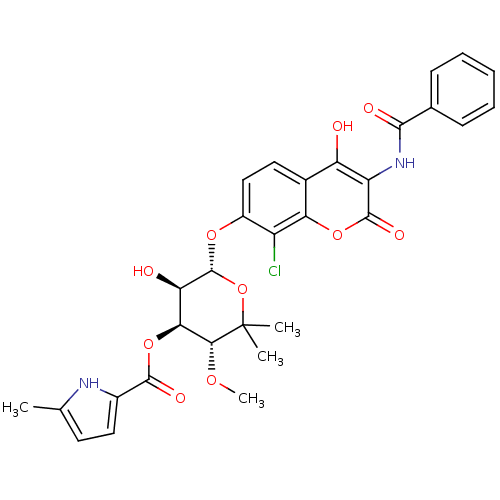 ((3R,4S,5R,6S)-6-(3-benzamido-8-chloro-4-hydroxy-2-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 72 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330316 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 73 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330317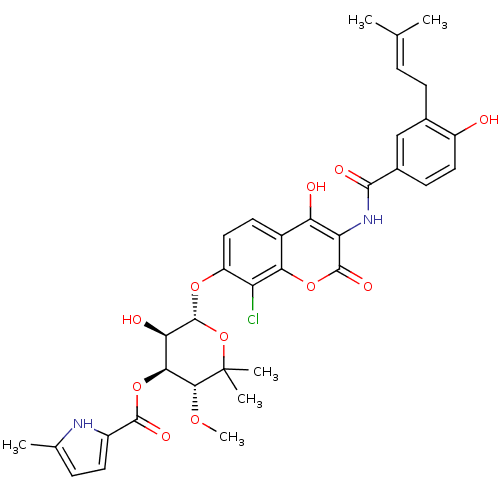 (5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | 73 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330305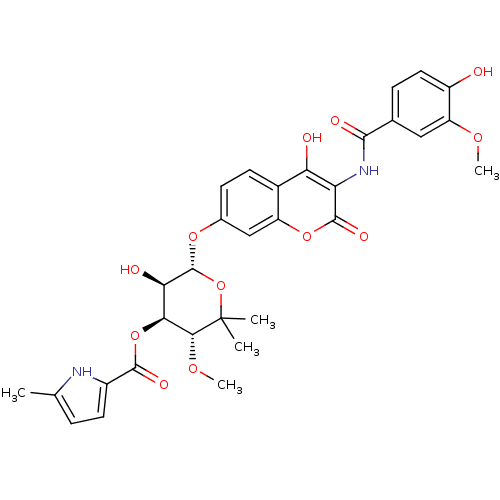 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 75 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50198322 (CHEMBL3936261) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 81 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli DNA gyrase B ATP binding site assessed as reduction in supercoiling of relaxed pNO1 DNA after 30 mins in presence of b... | J Med Chem 59: 8941-8954 (2016) Article DOI: 10.1021/acs.jmedchem.6b00864 BindingDB Entry DOI: 10.7270/Q25B04GJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330314 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-(methylth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 84 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330315 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 84 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50408190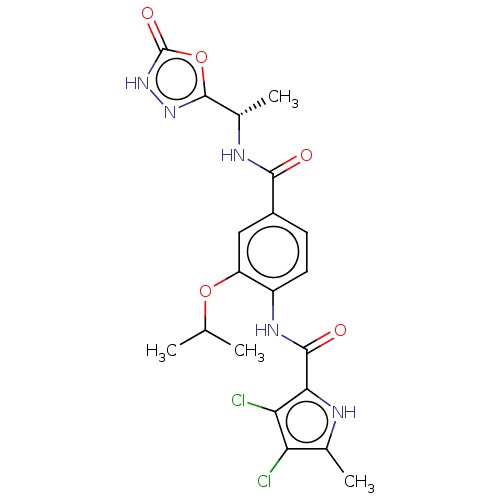 (CHEMBL4166687) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 85 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli C-terminal His6-tagged DNA gyrase B expressed in Escherichia coli BL21(DE3) preincubated for 10 mins followed by ATP a... | Eur J Med Chem 154: 117-132 (2018) Article DOI: 10.1016/j.ejmech.2018.05.011 BindingDB Entry DOI: 10.7270/Q2CV4M9N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330313 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 86 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50198235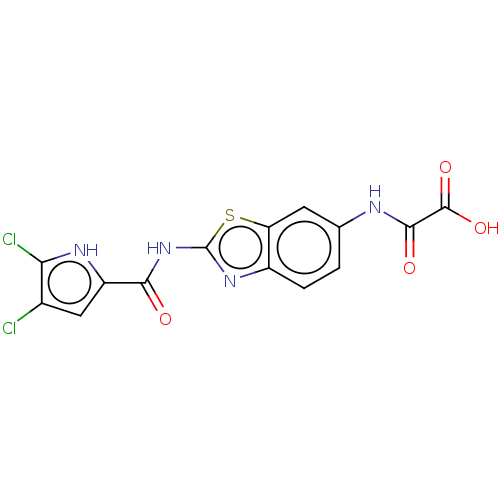 (CHEMBL3931016) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 87 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli DNA gyrase B ATP binding site assessed as reduction in supercoiling of relaxed pNO1 DNA after 30 mins in presence of b... | J Med Chem 59: 8941-8954 (2016) Article DOI: 10.1021/acs.jmedchem.6b00864 BindingDB Entry DOI: 10.7270/Q25B04GJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 149 total ) | Next | Last >> |