

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150768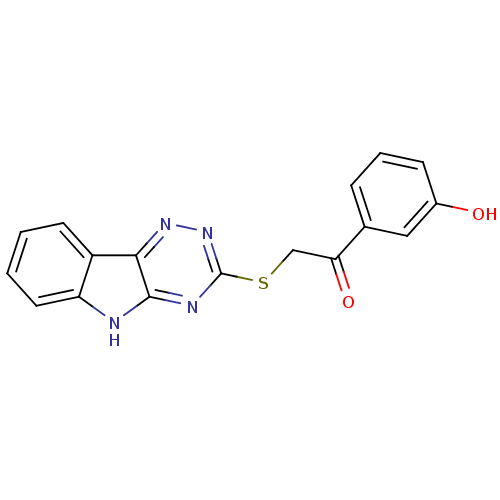 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-(3-h...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.46E+3 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150771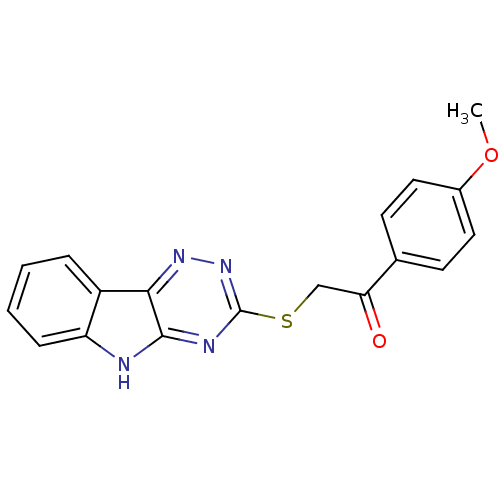 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-(4-m...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.89E+4 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150775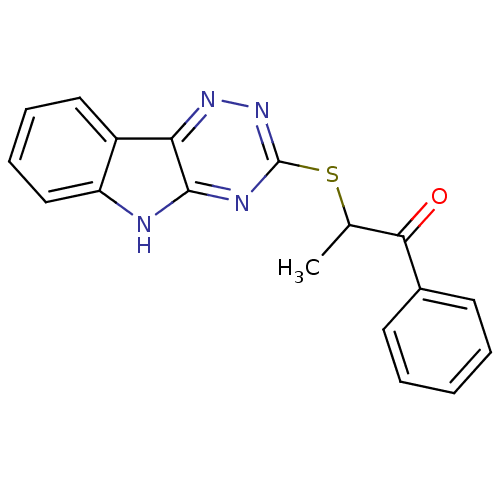 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-phen...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 3.69E+4 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150778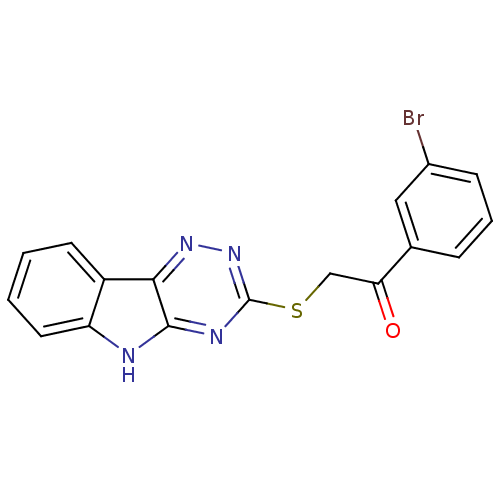 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-(3-b...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 3.71E+4 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150774 (3-(2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)acet...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 3.74E+4 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150770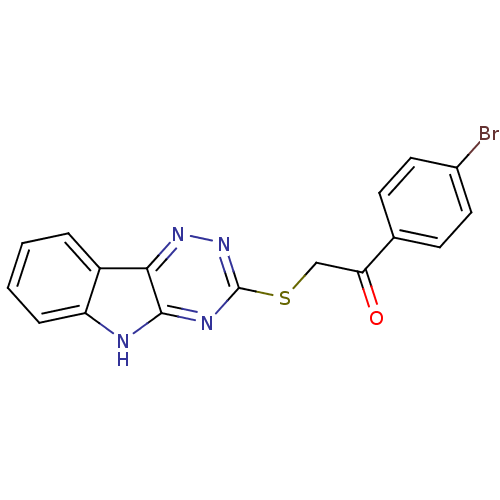 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-(4-b...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 3.78E+4 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150772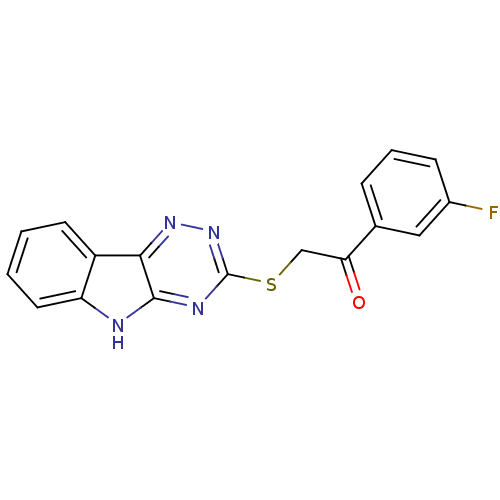 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-(3-f...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 3.81E+4 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM50333465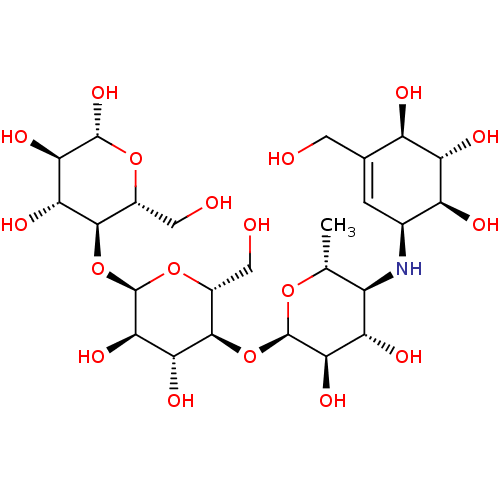 ((2R,3R,4R,5R,6R)-5-((2R,3R,4R,5S,6R)-5-((2R,3R,4S,...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 3.83E+4 | n/a | n/a | n/a | n/a | 6.8 | n/a |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150776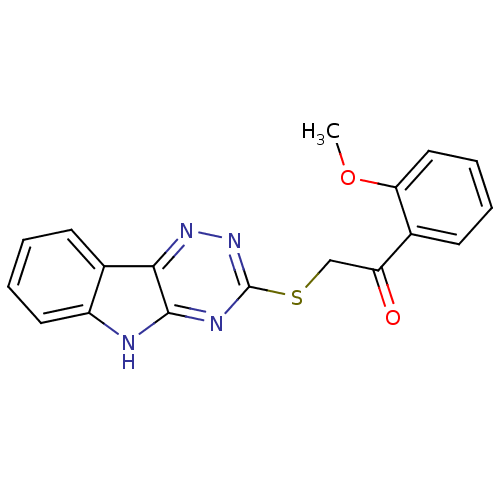 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-(2-m...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 5.16E+4 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150769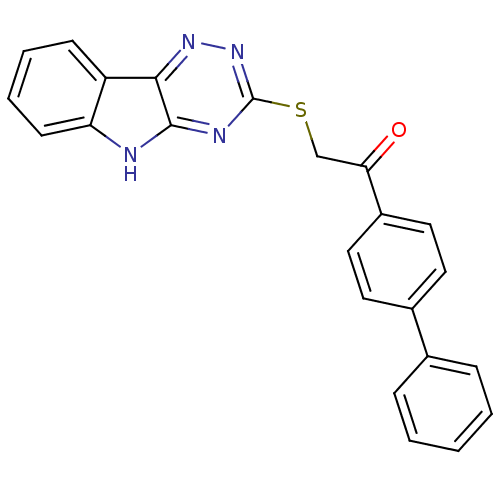 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-(bip...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 7.84E+4 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150773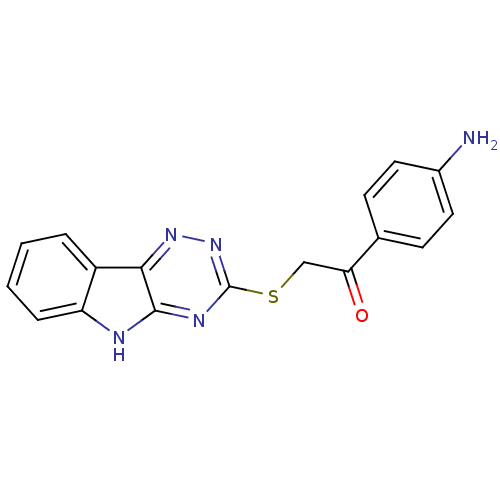 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-(4-n...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.25E+5 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Oligo-1,6-glucosidase IMA1 (Saccharomyces cerevisiae S288c (Baker's yeast)) | BDBM150777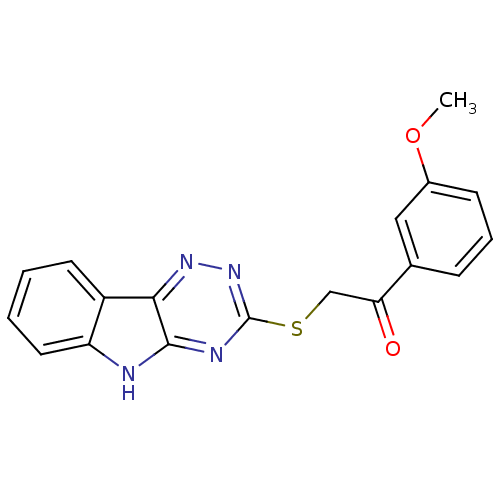 (2-(5H-[1,2,4]triazino[5,6-b]indol-3-ylthio)-1-(3-m...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 3.13E+5 | n/a | n/a | n/a | n/a | n/a | 310.15 |
Hazara University | Assay Description Typically, α-glucosidase activity was assayed in 50 mM phosphate buffer (pH 6.8) containing 5% v/v DMSO and PNP glycoside was used as a substrat... | Bioorg Chem 58: 81-7 (2015) Article DOI: 10.1016/j.bioorg.2014.12.001 BindingDB Entry DOI: 10.7270/Q2JQ0ZR8 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||