

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Endoplasmic reticulum chaperone BiP (382/382 = 100%)† (Homo sapiens (Human)) | BDBM82129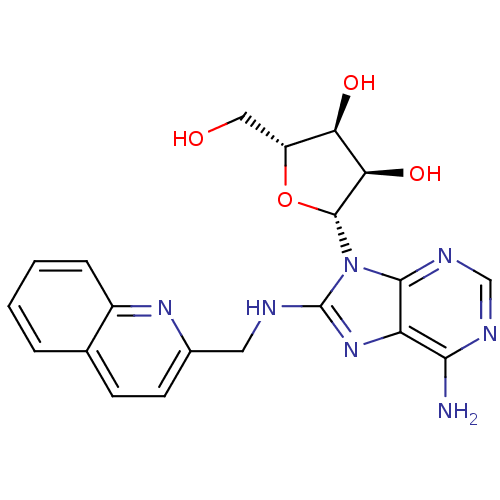 ((2R,3R,4S,5R)-2-{6-Amino-8-[(quinolin-2-ylmethyl)a...) | PDB KEGG UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 2.41E+3 | n/a | 0.400 | n/a | 7.4 | 25 |
Vernalis (R&D) Ltd. | Assay Description SPR measurements wereperformed on BIAcore T100 instrument (BIAcore GE Healthcare), at25 C on series S NTA chips (certified) according to provider'... | J Med Chem 54: 4034-41 (2011) Article DOI: 10.1021/jm101625x BindingDB Entry DOI: 10.7270/Q2R49P83 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||