

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15361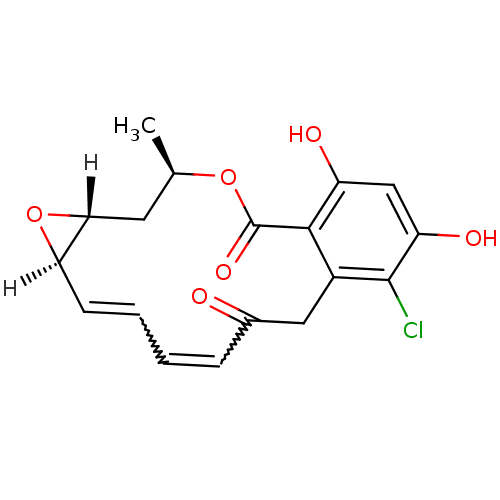 ((1aR,2Z,4E,14R,15aR)-8-chloro-9,11-dihydroxy-14-me...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | <200 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15384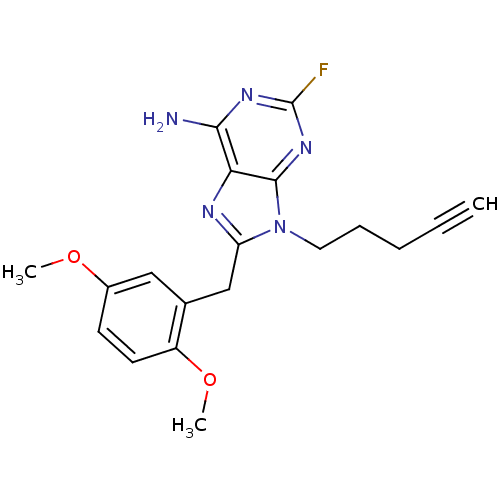 (8-(2,5-dimethoxybenzyl)-2-fluoro-9-pent-4-ynyl-9H-...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | 4.10E+3 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15359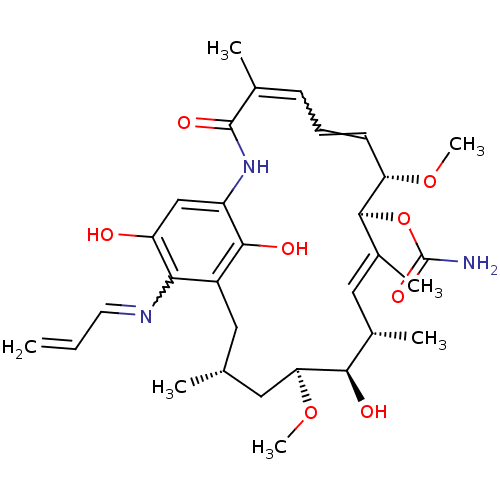 ((4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14-...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | 1.34E+4 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15383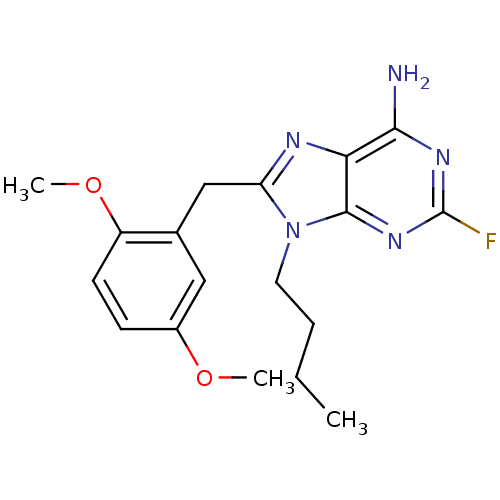 (9-butyl-8-(2,5-dimethoxybenzyl)-2-fluoro-9H-purin-...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 1.43E+4 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15377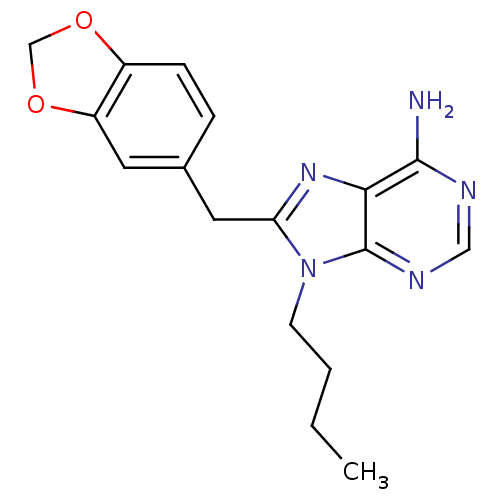 (8-(1,3-benzodioxol-5-ylmethyl)-9-butyl-9H-purin-6-...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 1.53E+4 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15385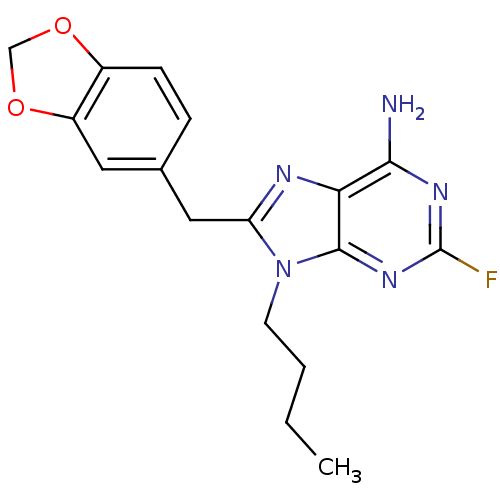 (8-(1,3-benzodioxol-5-ylmethyl)-9-butyl-2-fluoro-9H...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 1.71E+4 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15381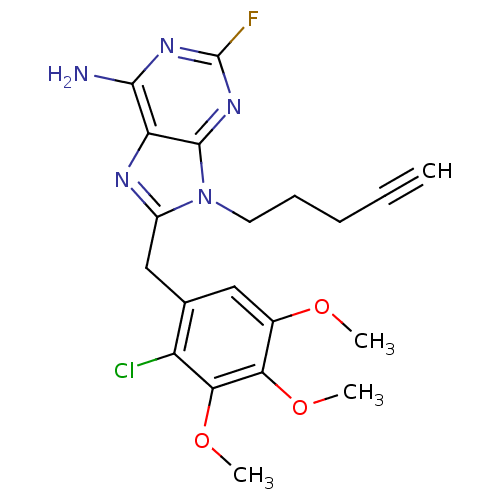 (8-(2-chloro-3,4,5-trimethoxybenzyl)-2-fluoro-9-pen...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | 3.00E+4 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15378 (9-butyl-8-(2,5-dimethoxybenzyl)-9H-purin-6-amine |...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | 4.10E+4 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15382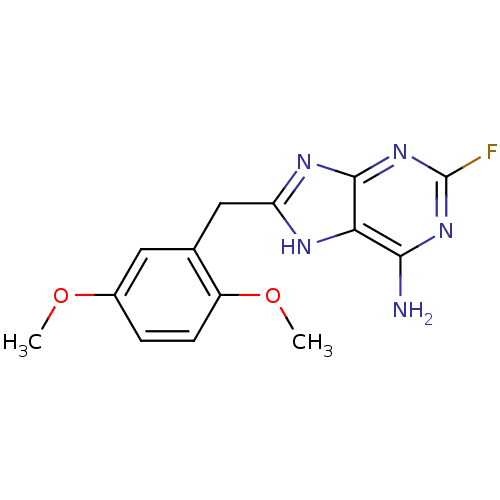 (8-(2,5-dimethoxybenzyl)-2-fluoro-7H-purin-6-amine ...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 5.35E+4 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15376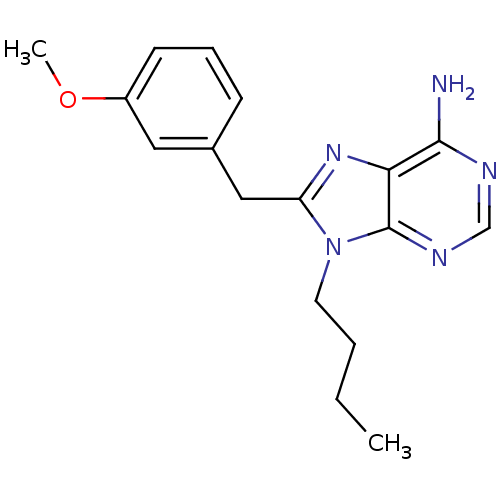 (9-butyl-8-(3-methoxybenzyl)-9H-purin-6-amine | 9-b...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | 7.50E+4 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15380 (8-(2-chloro-3,4,5-trimethoxybenzyl)-9-pent-4-ynyl-...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | >2.00E+5 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15375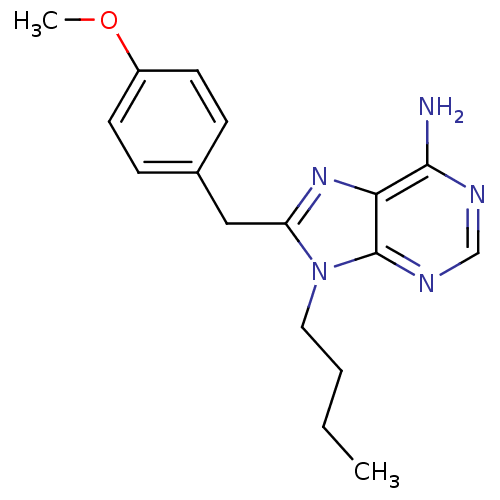 (9-butyl-8-(4-methoxybenzyl)-9H-purin-6-amine | 9-b...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | >2.00E+5 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15374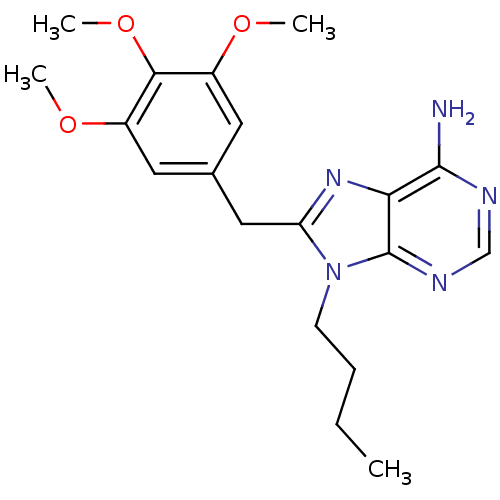 (9-butyl-8-(3,4,5-trimethoxybenzyl)-9H-purin-6-amin...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | >2.00E+5 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent molecular chaperone HSP82 (Saccharomyces cerevisiae) | BDBM15379 (9-butyl-8-(2-chloro-3,4,5-trimethoxybenzyl)-9H-pur...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | >2.00E+5 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Vernalis (R&D) Ltd | Assay Description The HSP90 ATPase activity was determined by following the procedure of malachite green assay. The assay is based on quantitation of the green complex... | Chem Biol 11: 775-85 (2004) Article DOI: 10.1016/j.chembiol.2004.03.033 BindingDB Entry DOI: 10.7270/Q2RX99BC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||