

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutamate racemase (Helicobacter pylori) | BDBM26431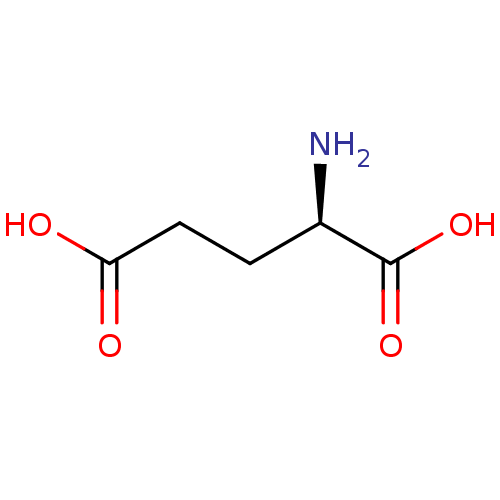 ((2R)-2-aminopentanedioic acid | CHEMBL76232 | D-Gl...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 5.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Global Structural Chemistry Curated by ChEMBL | Assay Description Inhibition of Helicobacter pylori glutamate racemase | Nature 447: 817-822 (2007) Article DOI: 10.1038/nature05689 BindingDB Entry DOI: 10.7270/Q2HX1DJ3 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate racemase (Helicobacter pylori) | BDBM50215445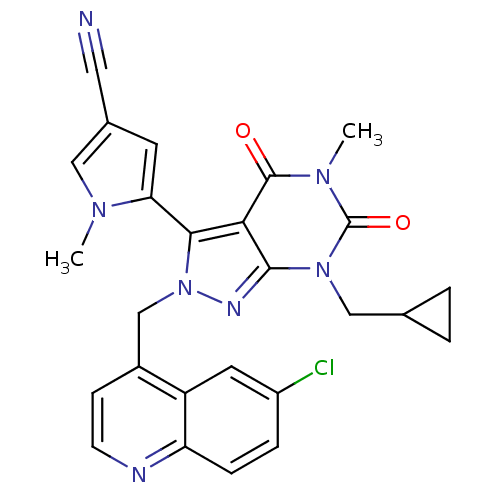 (5-(2-((6-chloroquinolin-4-yl)methyl)-7-(cyclopropy...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | 25 | n/a | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Global Structural Chemistry Curated by ChEMBL | Assay Description Inhibition of Helicobacter pylori glutamate racemase | Nature 447: 817-822 (2007) Article DOI: 10.1038/nature05689 BindingDB Entry DOI: 10.7270/Q2HX1DJ3 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate racemase (Helicobacter pylori) | BDBM50215444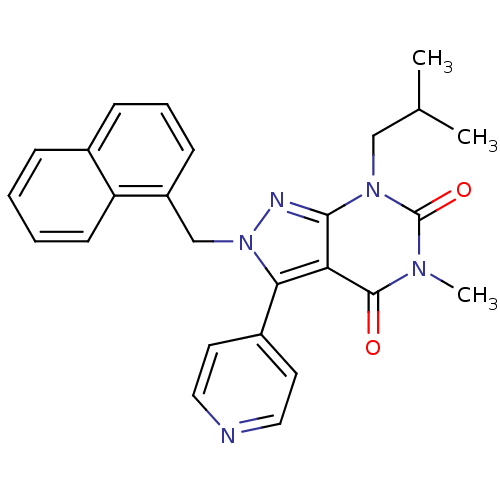 (5-METHYL-7-(2-METHYLPROPYL)-2-(NAPHTHALEN-1-YLMETH...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Patents | MMDB PDB Article PubMed | n/a | n/a | 1.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Global Structural Chemistry Curated by ChEMBL | Assay Description Inhibition of Helicobacter pylori glutamate racemase | Nature 447: 817-822 (2007) Article DOI: 10.1038/nature05689 BindingDB Entry DOI: 10.7270/Q2HX1DJ3 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate racemase (Streptococcus pneumoniae) | BDBM50215444 (5-METHYL-7-(2-METHYLPROPYL)-2-(NAPHTHALEN-1-YLMETH...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Patents | Article PubMed | n/a | n/a | >4.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Global Structural Chemistry Curated by ChEMBL | Assay Description Inhibition of MurI in Streptococcus pneumoniae | Nature 447: 817-822 (2007) Article DOI: 10.1038/nature05689 BindingDB Entry DOI: 10.7270/Q2HX1DJ3 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate racemase (Streptococcus pneumoniae) | BDBM50215445 (5-(2-((6-chloroquinolin-4-yl)methyl)-7-(cyclopropy...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | >4.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Global Structural Chemistry Curated by ChEMBL | Assay Description Inhibition of MurI in Streptococcus pneumoniae | Nature 447: 817-822 (2007) Article DOI: 10.1038/nature05689 BindingDB Entry DOI: 10.7270/Q2HX1DJ3 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate racemase (Helicobacter pylori) | BDBM50215444 (5-METHYL-7-(2-METHYLPROPYL)-2-(NAPHTHALEN-1-YLMETH...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Patents | MMDB PDB Article PubMed | n/a | n/a | n/a | 3.90E+3 | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Global Structural Chemistry Curated by ChEMBL | Assay Description Binding affinity to Helicobacter pylori MurI by protein fluorescence binding assay | Nature 447: 817-822 (2007) Article DOI: 10.1038/nature05689 BindingDB Entry DOI: 10.7270/Q2HX1DJ3 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate racemase (Helicobacter pylori) | BDBM50215445 (5-(2-((6-chloroquinolin-4-yl)methyl)-7-(cyclopropy...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | n/a | 25 | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Global Structural Chemistry Curated by ChEMBL | Assay Description Binding affinity to Helicobacter pylori MurI by protein fluorescence binding assay | Nature 447: 817-822 (2007) Article DOI: 10.1038/nature05689 BindingDB Entry DOI: 10.7270/Q2HX1DJ3 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate racemase (Helicobacter pylori) | BDBM50215444 (5-METHYL-7-(2-METHYLPROPYL)-2-(NAPHTHALEN-1-YLMETH...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MMDB PC cid PC sid PDB UniChem Patents | MMDB PDB Article PubMed | n/a | n/a | n/a | 2.90E+3 | n/a | n/a | n/a | n/a | n/a |
AstraZeneca Global Structural Chemistry Curated by ChEMBL | Assay Description Binding affinity to Helicobacter pylori MurI by isothermal titration calorimetry | Nature 447: 817-822 (2007) Article DOI: 10.1038/nature05689 BindingDB Entry DOI: 10.7270/Q2HX1DJ3 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||