 Found 7 hits of Enzyme Inhibition Constant Data
Found 7 hits of Enzyme Inhibition Constant Data Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
3-phosphoshikimate 1-carboxyvinyltransferase
(Escherichia coli (strain K12)) | BDBM50280240
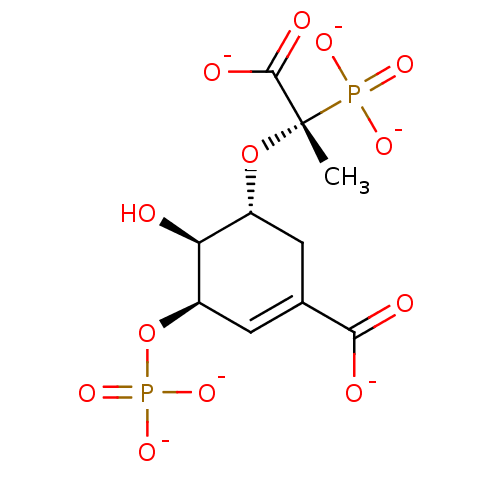
((3R,4S,5R)-4-hydroxy-5-[(1S)-1-methyl-2-oxido-2-ox...)Show SMILES C[C@@](O[C@@H]1CC(=C[C@@H](OP([O-])([O-])=O)[C@H]1O)C([O-])=O)(C([O-])=O)P([O-])([O-])=O |c:5| Show InChI InChI=1S/C10H16O13P2/c1-10(9(14)15,24(16,17)18)22-5-2-4(8(12)13)3-6(7(5)11)23-25(19,20)21/h3,5-7,11H,2H2,1H3,(H,12,13)(H,14,15)(H2,16,17,18)(H2,19,20,21)/p-6/t5-,6-,7+,10+/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
| 15 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Potency against EPSP synthase |
Bioorg Med Chem Lett 2: 1435-1440 (1992)
Article DOI: 10.1016/S0960-894X(00)80527-2
BindingDB Entry DOI: 10.7270/Q2XD1244 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
3-phosphoshikimate 1-carboxyvinyltransferase
(Escherichia coli (strain K12)) | BDBM50280239
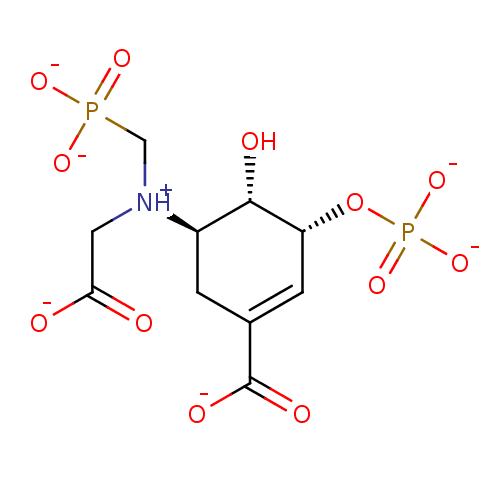
((3-Carboxy-6-hydroxy-5-phosphonooxy-cyclohex-3-eny...)Show SMILES O[C@@H]1[C@H](OP([O-])([O-])=O)C=C(C[C@H]1[NH+](CC([O-])=O)CP([O-])([O-])=O)C([O-])=O |c:8| Show InChI InChI=1S/C10H17NO12P2/c12-8(13)3-11(4-24(17,18)19)6-1-5(10(15)16)2-7(9(6)14)23-25(20,21)22/h2,6-7,9,14H,1,3-4H2,(H,12,13)(H,15,16)(H2,17,18,19)(H2,20,21,22)/p-5/t6-,7-,9+/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
| 7.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Inhibition of the EPSP synthase by the compound expressed as weak apparent value |
Bioorg Med Chem Lett 2: 1435-1440 (1992)
Article DOI: 10.1016/S0960-894X(00)80527-2
BindingDB Entry DOI: 10.7270/Q2XD1244 |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
3-phosphoshikimate 1-carboxyvinyltransferase
(Escherichia coli (strain K12)) | BDBM50280239

((3-Carboxy-6-hydroxy-5-phosphonooxy-cyclohex-3-eny...)Show SMILES O[C@@H]1[C@H](OP([O-])([O-])=O)C=C(C[C@H]1[NH+](CC([O-])=O)CP([O-])([O-])=O)C([O-])=O |c:8| Show InChI InChI=1S/C10H17NO12P2/c12-8(13)3-11(4-24(17,18)19)6-1-5(10(15)16)2-7(9(6)14)23-25(20,21)22/h2,6-7,9,14H,1,3-4H2,(H,12,13)(H,15,16)(H2,17,18,19)(H2,20,21,22)/p-5/t6-,7-,9+/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
| 1.30E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Dissociation constant of the at EPSP synthase was determined |
Bioorg Med Chem Lett 2: 1435-1440 (1992)
Article DOI: 10.1016/S0960-894X(00)80527-2
BindingDB Entry DOI: 10.7270/Q2XD1244 |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
3-phosphoshikimate 1-carboxyvinyltransferase
(Escherichia coli (strain K12)) | BDBM50280241
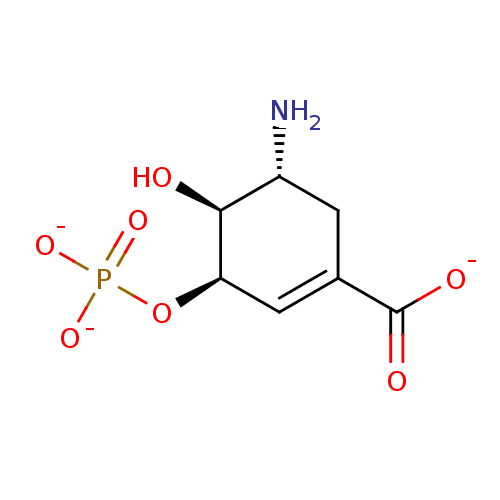
((3R,4S,5R)-5-amino-4-hydroxy-3-(phosphonatooxy)cyc...)Show SMILES N[C@@H]1CC(=C[C@@H](OP([O-])([O-])=O)[C@H]1O)C([O-])=O |c:3| Show InChI InChI=1S/C7H12NO7P/c8-4-1-3(7(10)11)2-5(6(4)9)15-16(12,13)14/h2,4-6,9H,1,8H2,(H,10,11)(H2,12,13,14)/p-3/t4-,5-,6+/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
| 8.50E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Potency against EPSP synthase |
Bioorg Med Chem Lett 2: 1435-1440 (1992)
Article DOI: 10.1016/S0960-894X(00)80527-2
BindingDB Entry DOI: 10.7270/Q2XD1244 |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
3-phosphoshikimate 1-carboxyvinyltransferase
(Escherichia coli (strain K12)) | BDBM50280239

((3-Carboxy-6-hydroxy-5-phosphonooxy-cyclohex-3-eny...)Show SMILES O[C@@H]1[C@H](OP([O-])([O-])=O)C=C(C[C@H]1[NH+](CC([O-])=O)CP([O-])([O-])=O)C([O-])=O |c:8| Show InChI InChI=1S/C10H17NO12P2/c12-8(13)3-11(4-24(17,18)19)6-1-5(10(15)16)2-7(9(6)14)23-25(20,21)22/h2,6-7,9,14H,1,3-4H2,(H,12,13)(H,15,16)(H2,17,18,19)(H2,20,21,22)/p-5/t6-,7-,9+/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
| n/a | n/a | n/a | 8.00E+3 | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Dissociation constant of the at EPSP synthase was determined |
Bioorg Med Chem Lett 2: 1435-1440 (1992)
Article DOI: 10.1016/S0960-894X(00)80527-2
BindingDB Entry DOI: 10.7270/Q2XD1244 |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
3-phosphoshikimate 1-carboxyvinyltransferase
(Escherichia coli (strain K12)) | BDBM50280242

((3R,4S,5R)-4,5-Dihydroxy-3-phosphonooxy-cyclohex-1...)Show SMILES O[C@@H]1CC(=C[C@@H](OP([O-])([O-])=O)[C@H]1O)C([O-])=O |c:3| Show InChI InChI=1S/C7H11O8P/c8-4-1-3(7(10)11)2-5(6(4)9)15-16(12,13)14/h2,4-6,8-9H,1H2,(H,10,11)(H2,12,13,14)/p-3/t4-,5-,6+/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| MCE
KEGG
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
| n/a | n/a | n/a | 8.00E+3 | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Dissociation constant of the at EPSP synthase was determined |
Bioorg Med Chem Lett 2: 1435-1440 (1992)
Article DOI: 10.1016/S0960-894X(00)80527-2
BindingDB Entry DOI: 10.7270/Q2XD1244 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
3-phosphoshikimate 1-carboxyvinyltransferase
(Escherichia coli (strain K12)) | BDBM50280240

((3R,4S,5R)-4-hydroxy-5-[(1S)-1-methyl-2-oxido-2-ox...)Show SMILES C[C@@](O[C@@H]1CC(=C[C@@H](OP([O-])([O-])=O)[C@H]1O)C([O-])=O)(C([O-])=O)P([O-])([O-])=O |c:5| Show InChI InChI=1S/C10H16O13P2/c1-10(9(14)15,24(16,17)18)22-5-2-4(8(12)13)3-6(7(5)11)23-25(19,20)21/h3,5-7,11H,2H2,1H3,(H,12,13)(H,14,15)(H2,16,17,18)(H2,19,20,21)/p-6/t5-,6-,7+,10+/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
| n/a | n/a | n/a | 15 | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Dissociation constant against EPSP synthase |
Bioorg Med Chem Lett 2: 1435-1440 (1992)
Article DOI: 10.1016/S0960-894X(00)80527-2
BindingDB Entry DOI: 10.7270/Q2XD1244 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data