| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM772 |
|---|
| Substrate/Competitor | BDBM12679 |
|---|
| Meas. Tech. | Enzyme Assay and Determination of the Inhibition Constants |
|---|
| pH | 7.4±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| Ki | 21000±2000 nM |
|---|
| Citation |  Katz, BA; Mackman, R; Luong, C; Radika, K; Martelli, A; Sprengeler, PA; Wang, J; Chan, H; Wong, L Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator. Chem Biol7:299-312 (2000) [PubMed] Article Katz, BA; Mackman, R; Luong, C; Radika, K; Martelli, A; Sprengeler, PA; Wang, J; Chan, H; Wong, L Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator. Chem Biol7:299-312 (2000) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 25790.52 |
|---|
| Organism: | Bos taurus (bovine) |
|---|
| Description: | P00760 |
|---|
| Residue: | 246 |
|---|
| Sequence: | MKTFIFLALLGAAVAFPVDDDDKIVGGYTCGANTVPYQVSLNSGYHFCGGSLINSQWVVS
AAHCYKSGIQVRLGEDNINVVEGNEQFISASKSIVHPSYNSNTLNNDIMLIKLKSAASLN
SRVASISLPTSCASAGTQCLISGWGNTKSSGTSYPDVLKCLKAPILSDSSCKSAYPGQIT
SNMFCAGYLEGGKDSCQGDSGGPVVCSGKLQGIVSWGSGCAQKNKPGVYTKVCNYVSWIK
QTIASN
|
|
|
|---|
| BDBM772 |
|---|
| BDBM12679 |
|---|
| Name | BDBM772 |
|---|
| Synonyms: | Benzamidine | CHEMBL79897 | [amino(phenyl)methylidene]azanium | benzamidine deriv. | benzamidinium chloride |
|---|
| Type | n/a |
|---|
| Emp. Form. | C7H9N2 |
|---|
| Mol. Mass. | 121.1592 |
|---|
| SMILES | NC(=[NH2+])c1ccccc1 |
|---|
| Structure | 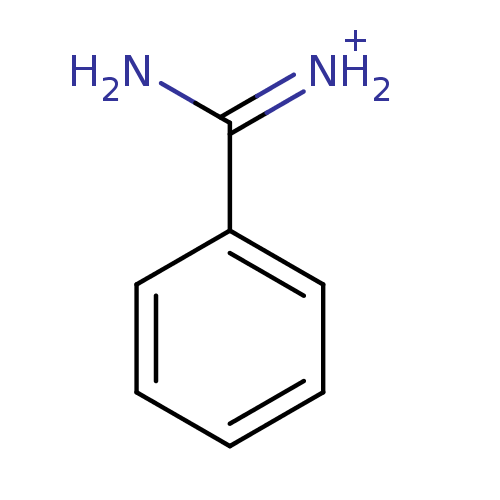
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Katz, BA; Mackman, R; Luong, C; Radika, K; Martelli, A; Sprengeler, PA; Wang, J; Chan, H; Wong, L Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator. Chem Biol7:299-312 (2000) [PubMed] Article
Katz, BA; Mackman, R; Luong, C; Radika, K; Martelli, A; Sprengeler, PA; Wang, J; Chan, H; Wong, L Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator. Chem Biol7:299-312 (2000) [PubMed] Article