

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Lysine-specific histone demethylase 1A [151-852] | ||
| Ligand | BDBM104112 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Demethylase Biochemical Assay | ||
| pH | 7.5±n/a | ||
| Temperature | 298.15±n/a K | ||
| IC50 | <50±n/a nM | ||
| Comments | extracted | ||
| Citation |  Wu, L; Konkol, LC; Lajkiewicz, N; Lu, L; Xu, M; Yao, W; Yu, Z; Zhang, C; He, C Substituted imidazo[1,5-α]pyridines and imidazo[1,5-α]pyrazines as LSD1 inhibitors US Patent US9695168 Publication Date 7/4/2017 Wu, L; Konkol, LC; Lajkiewicz, N; Lu, L; Xu, M; Yao, W; Yu, Z; Zhang, C; He, C Substituted imidazo[1,5-α]pyridines and imidazo[1,5-α]pyrazines as LSD1 inhibitors US Patent US9695168 Publication Date 7/4/2017 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Lysine-specific histone demethylase 1A [151-852] | |||
| Name: | Lysine-specific histone demethylase 1A [151-852] | ||
| Synonyms: | AOF2 | KDM1 | KDM1A | KDM1A_HUMAN | KIAA0601 | LSD1 | Lysine-specific histone demethylase 1A | ||
| Type: | n/a | ||
| Mol. Mass.: | 77867.68 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | ENZO (BML-SE544-0050) which consists of 151-852 aa | ||
| Residue: | 702 | ||
| Sequence: |
| ||
| BDBM104112 | |||
| n/a | |||
| Name | BDBM104112 | ||
| Synonyms: | US10640503, Example 30 | US9695168, 30 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H25FN6O3 | ||
| Mol. Mass. | 512.5349 | ||
| SMILES | CN1CCC[C@@H](COc2nc(-c3ccc(cc3)C#N)c(-c3cc4oc(=O)n(C)c4cc3F)n3cncc23)C1 |r,wD:5.5,(9.29,.9,;7.96,1.67,;7.96,3.21,;6.62,3.98,;5.29,3.21,;5.29,1.67,;3.96,.9,;2.62,1.67,;1.29,.9,;-.05,1.67,;-1.38,.9,;-2.71,1.67,;-2.71,3.21,;-4.05,3.98,;-5.43,3.24,;-5.38,1.67,;-4.05,.9,;-6.76,4.01,;-8.1,4.78,;-1.38,-.64,;-2.71,-1.41,;-4.05,-.64,;-5.38,-1.41,;-6.84,-.93,;-7.75,-2.18,;-9.29,-2.18,;-6.84,-3.42,;-7.61,-4.75,;-5.38,-2.95,;-4.05,-3.72,;-2.71,-2.95,;-1.38,-3.72,;-.05,-1.41,;.27,-2.91,;1.81,-3.07,;2.43,-1.67,;1.29,-.64,;6.62,.9,)| | ||
| Structure | 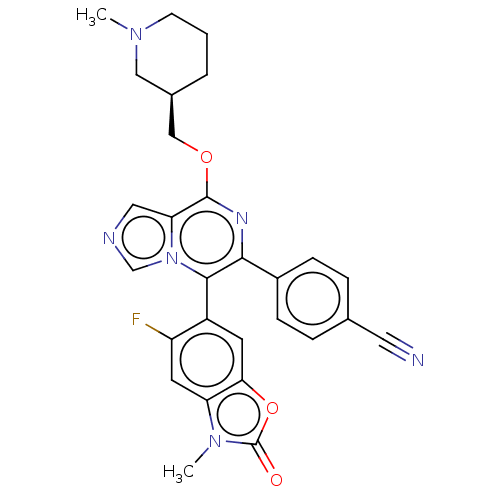
| ||