

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cyclin-dependent kinase 12 | ||
| Ligand | BDBM50367843 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1735959 (CHEMBL4151495) | ||
| IC50 | 460±n/a nM | ||
| Citation |  Ito, M; Tanaka, T; Toita, A; Uchiyama, N; Kokubo, H; Morishita, N; Klein, MG; Zou, H; Murakami, M; Kondo, M; Sameshima, T; Araki, S; Endo, S; Kawamoto, T; Morin, GB; Aparicio, SA; Nakanishi, A; Maezaki, H; Imaeda, Y Discovery of 3-Benzyl-1-( trans-4-((5-cyanopyridin-2-yl)amino)cyclohexyl)-1-arylurea Derivatives as Novel and Selective Cyclin-Dependent Kinase 12 (CDK12) Inhibitors. J Med Chem61:7710-7728 (2018) [PubMed] Article Ito, M; Tanaka, T; Toita, A; Uchiyama, N; Kokubo, H; Morishita, N; Klein, MG; Zou, H; Murakami, M; Kondo, M; Sameshima, T; Araki, S; Endo, S; Kawamoto, T; Morin, GB; Aparicio, SA; Nakanishi, A; Maezaki, H; Imaeda, Y Discovery of 3-Benzyl-1-( trans-4-((5-cyanopyridin-2-yl)amino)cyclohexyl)-1-arylurea Derivatives as Novel and Selective Cyclin-Dependent Kinase 12 (CDK12) Inhibitors. J Med Chem61:7710-7728 (2018) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cyclin-dependent kinase 12 | |||
| Name: | Cyclin-dependent kinase 12 | ||
| Synonyms: | 2.7.11.22 | 2.7.11.23 | CDC2-related protein kinase 7 | CDK12 | CDK12_HUMAN | CRK7 | CRKRS | Cdc2-related kinase, arginine/serine-rich | Cell division cycle 2-related protein kinase 7 | Cell division protein kinase 12 | Cyclin-dependent kinase 12 | KIAA0904 | hCDK12 | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 164218.64 | ||
| Organism: | Homo sapiens | ||
| Description: | ChEMBL_117739 | ||
| Residue: | 1490 | ||
| Sequence: |
| ||
| BDBM50367843 | |||
| n/a | |||
| Name | BDBM50367843 | ||
| Synonyms: | CHEMBL4174324 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H29N7O | ||
| Mol. Mass. | 443.5441 | ||
| SMILES | CCNC(=O)N([C@H]1CC[C@@H](CC1)Nc1ccc(cn1)C#N)c1ccc(cc1)-c1cnn(C)c1 |r,wU:6.5,wD:9.12,(20.34,-20.62,;21.68,-21.38,;23.01,-20.6,;24.35,-21.36,;25.68,-20.58,;24.28,-22.9,;22.96,-23.68,;22.97,-25.22,;21.64,-26,;20.3,-25.24,;20.29,-23.7,;21.61,-22.92,;18.97,-26.02,;17.63,-25.26,;16.3,-26.04,;14.96,-25.28,;14.95,-23.74,;16.28,-22.96,;17.62,-23.72,;13.61,-22.98,;12.27,-22.22,;25.58,-23.73,;26.95,-23.02,;28.25,-23.85,;28.18,-25.39,;26.81,-26.1,;25.52,-25.27,;29.48,-26.22,;29.57,-27.75,;31.07,-28.14,;31.89,-26.85,;33.43,-26.75,;30.92,-25.65,)| | ||
| Structure | 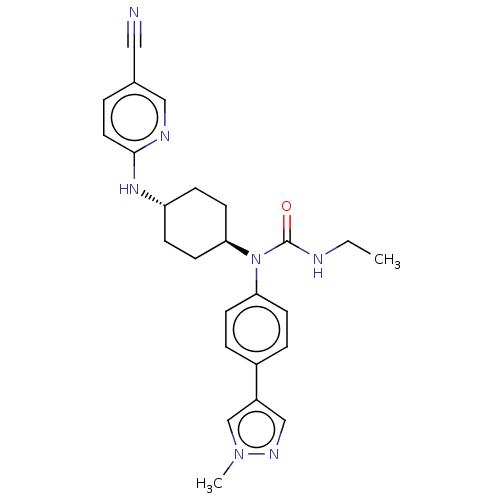
| ||