| Citation |  Kwak, SH; Shin, S; Lee, JH; Shim, JK; Kim, M; Lee, SD; Lee, A; Bae, J; Park, JH; Abdelrahman, A; Müller, CE; Cho, SK; Kang, SG; Bae, MA; Yang, JY; Ko, H; Goddard, WA; Kim, YC Synthesis and structure-activity relationships of quinolinone and quinoline-based P2X7 receptor antagonists and their anti-sphere formation activities in glioblastoma cells. Eur J Med Chem151:462-481 (2018) [PubMed] Article Kwak, SH; Shin, S; Lee, JH; Shim, JK; Kim, M; Lee, SD; Lee, A; Bae, J; Park, JH; Abdelrahman, A; Müller, CE; Cho, SK; Kang, SG; Bae, MA; Yang, JY; Ko, H; Goddard, WA; Kim, YC Synthesis and structure-activity relationships of quinolinone and quinoline-based P2X7 receptor antagonists and their anti-sphere formation activities in glioblastoma cells. Eur J Med Chem151:462-481 (2018) [PubMed] Article |
|---|
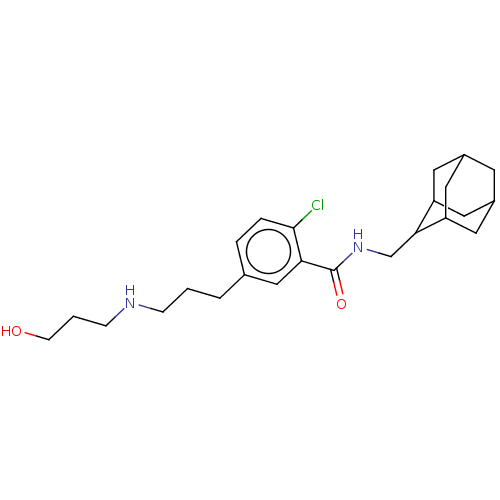
| SMILES | Cl.OCCCNCCCc1ccc(Cl)c(c1)C(=O)NCC1C2CC3CC(C2)CC1C3 |TLB:26:25:29:22.21.20,26:21:24.25.27:29,THB:20:21:24:27.28.29,20:28:24:22.26.21,19:20:24.25.27:29,(15.67,-31.77,;24.66,-28.77,;23.32,-28.01,;21.99,-28.78,;20.65,-28.02,;19.32,-28.79,;17.98,-28.03,;16.65,-28.8,;15.32,-28.04,;13.99,-28.81,;13.99,-30.36,;12.65,-31.13,;11.32,-30.36,;9.99,-31.13,;11.32,-28.82,;12.65,-28.05,;9.99,-28.05,;9.99,-26.51,;8.65,-28.82,;7.32,-28.05,;5.99,-28.82,;5.98,-30.35,;4.58,-30.69,;3.25,-30.2,;2.06,-31.48,;3.56,-31.06,;4.96,-31.63,;3.55,-29.48,;4.59,-28.24,;3.24,-28.72,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kwak, SH; Shin, S; Lee, JH; Shim, JK; Kim, M; Lee, SD; Lee, A; Bae, J; Park, JH; Abdelrahman, A; Müller, CE; Cho, SK; Kang, SG; Bae, MA; Yang, JY; Ko, H; Goddard, WA; Kim, YC Synthesis and structure-activity relationships of quinolinone and quinoline-based P2X7 receptor antagonists and their anti-sphere formation activities in glioblastoma cells. Eur J Med Chem151:462-481 (2018) [PubMed] Article
Kwak, SH; Shin, S; Lee, JH; Shim, JK; Kim, M; Lee, SD; Lee, A; Bae, J; Park, JH; Abdelrahman, A; Müller, CE; Cho, SK; Kang, SG; Bae, MA; Yang, JY; Ko, H; Goddard, WA; Kim, YC Synthesis and structure-activity relationships of quinolinone and quinoline-based P2X7 receptor antagonists and their anti-sphere formation activities in glioblastoma cells. Eur J Med Chem151:462-481 (2018) [PubMed] Article