| Reaction Details |
|---|
 | Report a problem with these data |
| Target | High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A |
|---|
| Ligand | BDBM50467436 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1796184 (CHEMBL4268301) |
|---|
| IC50 | 7760±n/a nM |
|---|
| Citation |  Ch?o?-Rzepa, G; ?lusarczyk, M; Jankowska, A; Gawalska, A; Bucki, A; Ko?aczkowski, M; ?wierczek, A; Pociecha, K; Wyska, E; Zygmunt, M; Kazek, G; Sa?at, K; Paw?owski, M Novel amide derivatives of 1,3-dimethyl-2,6-dioxopurin-7-yl-alkylcarboxylic acids as multifunctional TRPA1 antagonists and PDE4/7 inhibitors: A new approach for the treatment of pain. Eur J Med Chem158:517-533 (2018) [PubMed] Article Ch?o?-Rzepa, G; ?lusarczyk, M; Jankowska, A; Gawalska, A; Bucki, A; Ko?aczkowski, M; ?wierczek, A; Pociecha, K; Wyska, E; Zygmunt, M; Kazek, G; Sa?at, K; Paw?owski, M Novel amide derivatives of 1,3-dimethyl-2,6-dioxopurin-7-yl-alkylcarboxylic acids as multifunctional TRPA1 antagonists and PDE4/7 inhibitors: A new approach for the treatment of pain. Eur J Med Chem158:517-533 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A |
|---|
| Name: | High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | PDE7A | PDE7A_HUMAN | Phosphodiesterase 7 | Phosphodiesterase 7 (PDE7) | Phosphodiesterase 7A | Phosphodiesterase 7A (PDE7A1) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 55514.96 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13946 |
|---|
| Residue: | 482 |
|---|
| Sequence: | MEVCYQLPVLPLDRPVPQHVLSRRGAISFSSSSALFGCPNPRQLSQRRGAISYDSSDQTA
LYIRMLGDVRVRSRAGFESERRGSHPYIDFRIFHSQSEIEVSVSARNIRRLLSFQRYLRS
SRFFRGTAVSNSLNILDDDYNGQAKCMLEKVGNWNFDIFLFDRLTNGNSLVSLTFHLFSL
HGLIEYFHLDMMKLRRFLVMIQEDYHSQNPYHNAVHAADVTQAMHCYLKEPKLANSVTPW
DILLSLIAAATHDLDHPGVNQPFLIKTNHYLATLYKNTSVLENHHWRSAVGLLRESGLFS
HLPLESRQQMETQIGALILATDISRQNEYLSLFRSHLDRGDLCLEDTRHRHLVLQMALKC
ADICNPCRTWELSKQWSEKVTEEFFHQGDIEKKYHLGVSPLCDRHTESIANIQIGFMTYL
VEPLFTEWARFSNTRLSQTMLGHVGLNKASWKGLQREQSSSEDTDAAFELNSQLLPQENR
LS
|
|
|
|---|
| BDBM50467436 |
|---|
| n/a |
|---|
| Name | BDBM50467436 |
|---|
| Synonyms: | CHEMBL4284497 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H33N5O4 |
|---|
| Mol. Mass. | 455.5499 |
|---|
| SMILES | CCCOc1nc2n(C)c(=O)n(C)c(=O)c2n1CCCC(=O)Nc1ccc(cc1)C(C)(C)C |
|---|
| Structure | 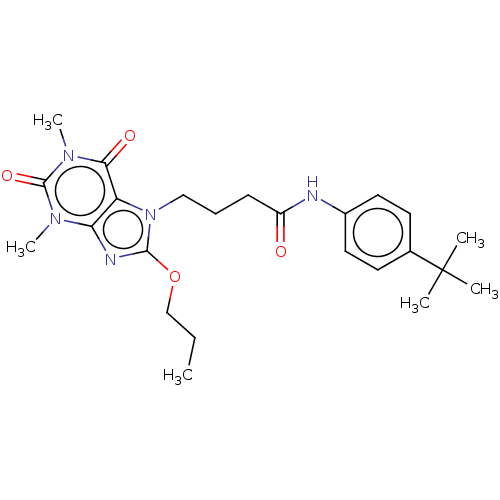
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ch?o?-Rzepa, G; ?lusarczyk, M; Jankowska, A; Gawalska, A; Bucki, A; Ko?aczkowski, M; ?wierczek, A; Pociecha, K; Wyska, E; Zygmunt, M; Kazek, G; Sa?at, K; Paw?owski, M Novel amide derivatives of 1,3-dimethyl-2,6-dioxopurin-7-yl-alkylcarboxylic acids as multifunctional TRPA1 antagonists and PDE4/7 inhibitors: A new approach for the treatment of pain. Eur J Med Chem158:517-533 (2018) [PubMed] Article
Ch?o?-Rzepa, G; ?lusarczyk, M; Jankowska, A; Gawalska, A; Bucki, A; Ko?aczkowski, M; ?wierczek, A; Pociecha, K; Wyska, E; Zygmunt, M; Kazek, G; Sa?at, K; Paw?owski, M Novel amide derivatives of 1,3-dimethyl-2,6-dioxopurin-7-yl-alkylcarboxylic acids as multifunctional TRPA1 antagonists and PDE4/7 inhibitors: A new approach for the treatment of pain. Eur J Med Chem158:517-533 (2018) [PubMed] Article