| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 3 |
|---|
| Ligand | BDBM50258579 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2109868 (CHEMBL4818543) |
|---|
| IC50 | 0.600000±n/a nM |
|---|
| Citation |  Beshore, DC; Adam, GC; Barnard, RJO; Burlein, C; Gallicchio, SN; Holloway, MK; Krosky, D; Lemaire, W; Myers, RW; Patel, S; Plotkin, MA; Powell, DA; Rada, V; Cox, CD; Coleman, PJ; Klein, DJ; Wolkenberg, SE Redefining the Histone Deacetylase Inhibitor Pharmacophore: High Potency with No Zinc Cofactor Interaction. ACS Med Chem Lett12:540-547 (2021) [PubMed] Article Beshore, DC; Adam, GC; Barnard, RJO; Burlein, C; Gallicchio, SN; Holloway, MK; Krosky, D; Lemaire, W; Myers, RW; Patel, S; Plotkin, MA; Powell, DA; Rada, V; Cox, CD; Coleman, PJ; Klein, DJ; Wolkenberg, SE Redefining the Histone Deacetylase Inhibitor Pharmacophore: High Potency with No Zinc Cofactor Interaction. ACS Med Chem Lett12:540-547 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 3 |
|---|
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | HD3 | HDAC3 | HDAC3_HUMAN | Histone deacetylase 3 (HDAC3) | Human HDAC3 | RPD3-2 | SMAP45 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48829.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15379 |
|---|
| Residue: | 428 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
NQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKESDVEI
|
|
|
|---|
| BDBM50258579 |
|---|
| n/a |
|---|
| Name | BDBM50258579 |
|---|
| Synonyms: | (S)-N-(1-(5-(2-methoxyquinolin-3-yl)-1H-imidazol-2-yl)-7-oxooctyl)-1-methylazetidine-3-carboxamide | CHEMBL2448576 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H35N5O3 |
|---|
| Mol. Mass. | 477.5985 |
|---|
| SMILES | CCC(=O)CCCCC[C@H](NC(=O)C1CN(C)C1)c1ncc([nH]1)-c1cc2ccccc2nc1OC |r| |
|---|
| Structure | 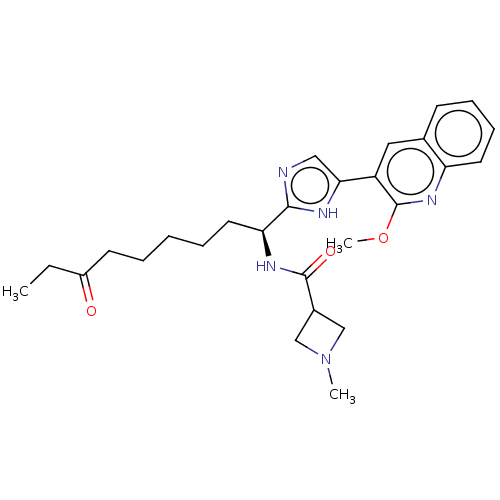
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Beshore, DC; Adam, GC; Barnard, RJO; Burlein, C; Gallicchio, SN; Holloway, MK; Krosky, D; Lemaire, W; Myers, RW; Patel, S; Plotkin, MA; Powell, DA; Rada, V; Cox, CD; Coleman, PJ; Klein, DJ; Wolkenberg, SE Redefining the Histone Deacetylase Inhibitor Pharmacophore: High Potency with No Zinc Cofactor Interaction. ACS Med Chem Lett12:540-547 (2021) [PubMed] Article
Beshore, DC; Adam, GC; Barnard, RJO; Burlein, C; Gallicchio, SN; Holloway, MK; Krosky, D; Lemaire, W; Myers, RW; Patel, S; Plotkin, MA; Powell, DA; Rada, V; Cox, CD; Coleman, PJ; Klein, DJ; Wolkenberg, SE Redefining the Histone Deacetylase Inhibitor Pharmacophore: High Potency with No Zinc Cofactor Interaction. ACS Med Chem Lett12:540-547 (2021) [PubMed] Article