| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform |
|---|
| Ligand | BDBM25028 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2225466 (CHEMBL5138979) |
|---|
| IC50 | 52±n/a nM |
|---|
| Citation |  Fairhurst, RA; Furet, P; Imbach-Weese, P; Stauffer, F; Rueeger, H; McCarthy, C; Ripoche, S; Oswald, S; Arnaud, B; Jary, A; Maira, M; Schnell, C; Guthy, DA; Wartmann, M; Kiffe, M; Desrayaud, S; Blasco, F; Widmer, T; Seiler, F; Gutmann, S; Knapp, M; Caravatti, G Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor. J Med Chem65:8345-8379 (2022) [PubMed] Article Fairhurst, RA; Furet, P; Imbach-Weese, P; Stauffer, F; Rueeger, H; McCarthy, C; Ripoche, S; Oswald, S; Arnaud, B; Jary, A; Maira, M; Schnell, C; Guthy, DA; Wartmann, M; Kiffe, M; Desrayaud, S; Blasco, F; Widmer, T; Seiler, F; Gutmann, S; Knapp, M; Caravatti, G Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor. J Med Chem65:8345-8379 (2022) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform |
|---|
| Name: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform |
|---|
| Synonyms: | PI3-kinase subunit alpha | PI3K-alpha | PI3Kalpha | PK3CA_RAT | Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit alpha | Phosphoinositide-3-kinase catalytic alpha polypeptide | Pik3ca | PtdIns-3-kinase subunit alpha | PtdIns-3-kinase subunit p110-alpha | Serine/threonine protein kinase PIK3CA | p110alpha |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 124363.80 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | A0A0G2K344 |
|---|
| Residue: | 1068 |
|---|
| Sequence: | MPPRPSSGELWGIHLMPPRILVECLLPNGMIVTLECLREATLVTIKHELFKEARKYPLHQ
LLQDESSYIFVSVTQEAEREEFFDETRRLCDLRLFQPFLKVIEPVGNREEKILNREIGFV
IGMPVCEFDMVKDPEVQDFRRNILNVCKEAVDLRDLNSPHSRAMYVYPPNVESSPELPKH
IYNKLDKGQIIVVIWVIVSPNNDKQKYTLKINHDCVPEQVIAEAIRKKTRSMLLSSEQLK
LCVLEYQGKYILKVCGCDEYFLEKYPLSQYKYIRSCIMLGRMPNLMLMAKESLYSQLPID
SFTMPSYSRRISTATPYMNGETATKSLWVINSALRIKILCATYVNVNIRDIDKIYVRTGI
YHGGEPLCDNVNTQRVPCSNPRWNEWLNYDIYIPDLPRAARLCLSICSVKGRKGAKEEHC
PLAWGNINLFDYTDTLVSGKMALNLWPVPHGLEDLLNPIGVTGSNPNKETPCLELEFDWF
SSVVKFPDMSVIEEHANWSVSREAGFSYSHTGLSNRLARDNELRENDKEQLRALCTRDPL
SEITEQEKDFLWSHRHYCVTIPEILPKLLLSVKWNSRDEVAQMYCLVKDWPPIKPEQAME
LLDCNYPDPMVRSFAVRCLEKYLTDDKLSQYLIQLVQVLKYEQYLDNLLVRFLLKKALTN
QRIGHFFFWHLKSEMHNKTVSQRFGLLLESYCRACGMYLKHLNRQVEAMEKLINLTDILK
QEKKDETQKVQMKFLVEQMRQPDFMDALQGFLSPLNPAHQLGNLRLEECRIMSSAKRPLW
LNWENPDIMSELLFQNNEIIFKNGDDLRQDMLTLQIIRIMENIWQNQGLDLRMLPYGCLS
IGDCVGLIEVVRNSHTIMQIQCKGGLKGALQFNSHTLHQWLKDKNKGEIYDAAIDLFTRS
CAGYCVATFILGIGDRHNSNIMVKDDGQLFHIDFGHFLDHKKKKFGYKRERVPFVLTQDF
LIVISKGAQEYTKTREFERFQEMCYKAYLAIRQHANLFINLFSMMLGSGMPELQSFDDIA
YIRKTLALDKTEQEALEYFTKQMNDAHHGGWTTKMDWIFHTIKQHALN
|
|
|
|---|
| BDBM25028 |
|---|
| n/a |
|---|
| Name | BDBM25028 |
|---|
| Synonyms: | 4-[2-(1H-indazol-4-yl)-6-[(4-methanesulfonylpiperazin-1-yl)methyl]thieno[3,2-d]pyrimidin-4-yl]morpholine | GDC-0941 | GDC0941 | US10112932, pictilisib | US10851091, Compound pictilisib |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H27N7O3S2 |
|---|
| Mol. Mass. | 513.636 |
|---|
| SMILES | CS(=O)(=O)N1CCN(Cc2cc3nc(nc(N4CCOCC4)c3s2)-c2cccc3[nH]ncc23)CC1 |
|---|
| Structure | 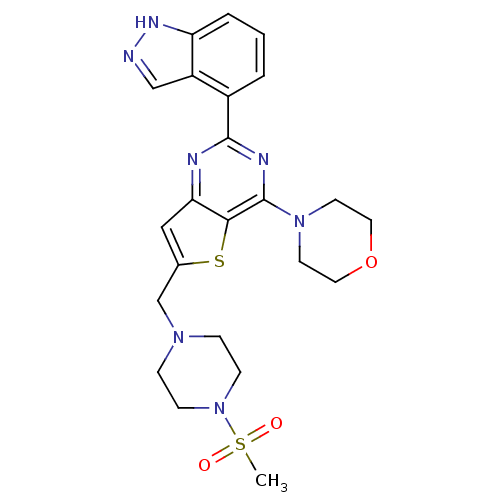
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Fairhurst, RA; Furet, P; Imbach-Weese, P; Stauffer, F; Rueeger, H; McCarthy, C; Ripoche, S; Oswald, S; Arnaud, B; Jary, A; Maira, M; Schnell, C; Guthy, DA; Wartmann, M; Kiffe, M; Desrayaud, S; Blasco, F; Widmer, T; Seiler, F; Gutmann, S; Knapp, M; Caravatti, G Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor. J Med Chem65:8345-8379 (2022) [PubMed] Article
Fairhurst, RA; Furet, P; Imbach-Weese, P; Stauffer, F; Rueeger, H; McCarthy, C; Ripoche, S; Oswald, S; Arnaud, B; Jary, A; Maira, M; Schnell, C; Guthy, DA; Wartmann, M; Kiffe, M; Desrayaud, S; Blasco, F; Widmer, T; Seiler, F; Gutmann, S; Knapp, M; Caravatti, G Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor. J Med Chem65:8345-8379 (2022) [PubMed] Article