| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Integrase |
|---|
| Ligand | BDBM50187187 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_383533 (CHEMBL867996) |
|---|
| IC50 | 255±n/a nM |
|---|
| Citation |  Fardis, M; Jin, H; Jabri, S; Cai, RZ; Mish, M; Tsiang, M; Kim, CU Effect of substitution on novel tricyclic HIV-1 integrase inhibitors. Bioorg Med Chem Lett16:4031-5 (2006) [PubMed] Article Fardis, M; Jin, H; Jabri, S; Cai, RZ; Mish, M; Tsiang, M; Kim, CU Effect of substitution on novel tricyclic HIV-1 integrase inhibitors. Bioorg Med Chem Lett16:4031-5 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Integrase |
|---|
| Name: | Integrase |
|---|
| Synonyms: | Human immunodeficiency virus type 1 integrase |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 32231.48 |
|---|
| Organism: | Human immunodeficiency virus 1 |
|---|
| Description: | ChEMBL_90865 |
|---|
| Residue: | 288 |
|---|
| Sequence: | FLDGIDKAQDEHEKYHSNWRAMASDFNLPPVVAKEIVASCDKCQLKGEAMHGQVDCSPGI
WQLDCTHLEGKVILVAVHVASGYIEAEVIPAETGQETAYFLLKLAGRWPVKTIHTDNGSN
FTSTTVKAACWWAGIKQEFGIPYNPQSQGVVESMNKELKKIIGQVRDQAEHLKTAVQMAV
FIHNFKRKGGIGGYSAGERIVDIIATDIQTKELQKQITKIQNFRVYYRDSRDPLWKGPAK
LLWKGEGAVVIQDNSDIKVVPRRKVKIIRDYGKQMAGDDCVASRQDED
|
|
|
|---|
| BDBM50187187 |
|---|
| n/a |
|---|
| Name | BDBM50187187 |
|---|
| Synonyms: | 7-(4-fluorobenzyl)-9-hydroxy-8-oxo-7,8-dihydro-6H-pyrrolo[3,4-g]quinolin-5-yl dimethylsulfamate | CHEMBL214371 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H18FN3O5S |
|---|
| Mol. Mass. | 431.437 |
|---|
| SMILES | CN(C)S(=O)(=O)Oc1c2CN(Cc3ccc(F)cc3)C(=O)c2c(O)c2ncccc12 |
|---|
| Structure | 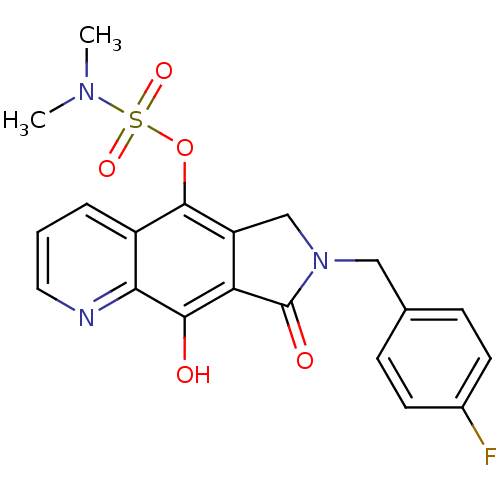
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Fardis, M; Jin, H; Jabri, S; Cai, RZ; Mish, M; Tsiang, M; Kim, CU Effect of substitution on novel tricyclic HIV-1 integrase inhibitors. Bioorg Med Chem Lett16:4031-5 (2006) [PubMed] Article
Fardis, M; Jin, H; Jabri, S; Cai, RZ; Mish, M; Tsiang, M; Kim, CU Effect of substitution on novel tricyclic HIV-1 integrase inhibitors. Bioorg Med Chem Lett16:4031-5 (2006) [PubMed] Article