| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Matrix metalloproteinase-14 |
|---|
| Ligand | BDBM50198033 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_432509 (CHEMBL918803) |
|---|
| IC50 | 11±n/a nM |
|---|
| Citation |  Biasone, A; Tortorella, P; Campestre, C; Agamennone, M; Preziuso, S; Chiappini, M; Nuti, E; Carelli, P; Rossello, A; Mazza, F; Gallina, C alpha-Biphenylsulfonylamino 2-methylpropyl phosphonates: enantioselective synthesis and selective inhibition of MMPs. Bioorg Med Chem15:791-9 (2006) [PubMed] Article Biasone, A; Tortorella, P; Campestre, C; Agamennone, M; Preziuso, S; Chiappini, M; Nuti, E; Carelli, P; Rossello, A; Mazza, F; Gallina, C alpha-Biphenylsulfonylamino 2-methylpropyl phosphonates: enantioselective synthesis and selective inhibition of MMPs. Bioorg Med Chem15:791-9 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Matrix metalloproteinase-14 |
|---|
| Name: | Matrix metalloproteinase-14 |
|---|
| Synonyms: | MMP-14 | MMP-X1 | MMP14 | MMP14_HUMAN | MT-MMP 1 | MT1-MMP | MT1MMP | MTMMP1 | Matrix Metalloproteinase-14 (MMP-14) | Matrix metalloproteinase 14 | Matrix metalloproteinase-14 | Matrix metalloproteinase-14 (MMP14) | Membrane-type matrix metalloproteinase 1 | Membrane-type-1 matrix metalloproteinase |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 65900.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P50281 |
|---|
| Residue: | 582 |
|---|
| Sequence: | MSPAPRPPRCLLLPLLTLGTALASLGSAQSSSFSPEAWLQQYGYLPPGDLRTHTQRSPQS
LSAAIAAMQKFYGLQVTGKADADTMKAMRRPRCGVPDKFGAEIKANVRRKRYAIQGLKWQ
HNEITFCIQNYTPKVGEYATYEAIRKAFRVWESATPLRFREVPYAYIREGHEKQADIMIF
FAEGFHGDSTPFDGEGGFLAHAYFPGPNIGGDTHFDSAEPWTVRNEDLNGNDIFLVAVHE
LGHALGLEHSSDPSAIMAPFYQWMDTENFVLPDDDRRGIQQLYGGESGFPTKMPPQPRTT
SRPSVPDKPKNPTYGPNICDGNFDTVAMLRGEMFVFKERWFWRVRNNQVMDGYPMPIGQF
WRGLPASINTAYERKDGKFVFFKGDKHWVFDEASLEPGYPKHIKELGRGLPTDKIDAALF
WMPNGKTYFFRGNKYYRFNEELRAVDSEYPKNIKVWEGIPESPRGSFMGSDEVFTYFYKG
NKYWKFNNQKLKVEPGYPKSALRDWMGCPSGGRPDEGTEEETEVIIIEVDEEGGGAVSAA
AVVLPVLLLLLVLAVGLAVFFFRRHGTPRRLLYCQRSLLDKV
|
|
|
|---|
| BDBM50198033 |
|---|
| n/a |
|---|
| Name | BDBM50198033 |
|---|
| Synonyms: | CHEMBL227484 | [(R)-2-methyl-1-(4'-methyl-biphenyl-4-sulfonylamino)-propyl]-phosphonic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H22NO5PS |
|---|
| Mol. Mass. | 383.399 |
|---|
| SMILES | CC(C)[C@H](NS(=O)(=O)c1ccc(cc1)-c1ccc(C)cc1)P(O)(O)=O |
|---|
| Structure | 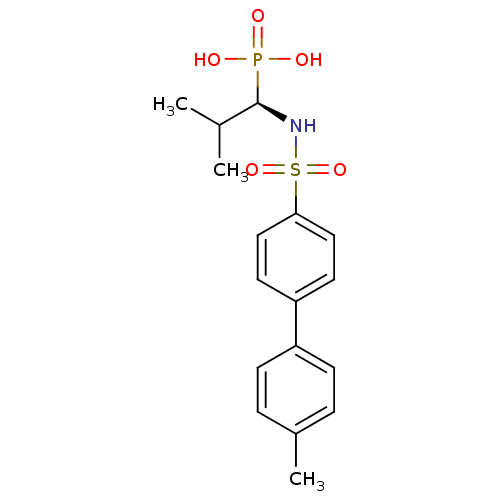
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Biasone, A; Tortorella, P; Campestre, C; Agamennone, M; Preziuso, S; Chiappini, M; Nuti, E; Carelli, P; Rossello, A; Mazza, F; Gallina, C alpha-Biphenylsulfonylamino 2-methylpropyl phosphonates: enantioselective synthesis and selective inhibition of MMPs. Bioorg Med Chem15:791-9 (2006) [PubMed] Article
Biasone, A; Tortorella, P; Campestre, C; Agamennone, M; Preziuso, S; Chiappini, M; Nuti, E; Carelli, P; Rossello, A; Mazza, F; Gallina, C alpha-Biphenylsulfonylamino 2-methylpropyl phosphonates: enantioselective synthesis and selective inhibition of MMPs. Bioorg Med Chem15:791-9 (2006) [PubMed] Article