

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 2D6 | ||
| Ligand | BDBM50329317 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_675409 (CHEMBL1273489) | ||
| IC50 | 30000±n/a nM | ||
| Citation |  Tice, CM; Zhao, W; Krosky, PM; Kruk, BA; Berbaum, J; Johnson, JA; Bukhtiyarov, Y; Panemangalore, R; Scott, BB; Zhao, Y; Bruno, JG; Howard, L; Togias, J; Ye, YJ; Singh, SB; McKeever, BM; Lindblom, PR; Guo, J; Guo, R; Nar, H; Schuler-Metz, A; Gregg, RE; Leftheris, K; Harrison, RK; McGeehan, GM; Zhuang, L; Claremon, DA Discovery and optimization of adamantyl carbamate inhibitors of 11ß-HSD1. Bioorg Med Chem Lett20:6725-9 (2010) [PubMed] Article Tice, CM; Zhao, W; Krosky, PM; Kruk, BA; Berbaum, J; Johnson, JA; Bukhtiyarov, Y; Panemangalore, R; Scott, BB; Zhao, Y; Bruno, JG; Howard, L; Togias, J; Ye, YJ; Singh, SB; McKeever, BM; Lindblom, PR; Guo, J; Guo, R; Nar, H; Schuler-Metz, A; Gregg, RE; Leftheris, K; Harrison, RK; McGeehan, GM; Zhuang, L; Claremon, DA Discovery and optimization of adamantyl carbamate inhibitors of 11ß-HSD1. Bioorg Med Chem Lett20:6725-9 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 2D6 | |||
| Name: | Cytochrome P450 2D6 | ||
| Synonyms: | CP2D6_HUMAN | CYP2D6 | CYP2DL1 | CYPIID6 | Cytochrome P450 2D6 (CYP2D6) | Debrisoquine 4-hydroxylase | P450-DB1 | ||
| Type: | Protein | ||
| Mol. Mass.: | 55774.82 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P10635 | ||
| Residue: | 497 | ||
| Sequence: |
| ||
| BDBM50329317 | |||
| n/a | |||
| Name | BDBM50329317 | ||
| Synonyms: | (S)-3-(3-Cyano-pyridin-2-ylamino)-pyrrolidine-1-carboxylic acid 5-carbamoyl-adamantan-2-yl ester | CHEMBL1270002 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H27N5O3 | ||
| Mol. Mass. | 409.4815 | ||
| SMILES | NC(=O)[C@@]12CC3CC(C1)[C@H](OC(=O)N1CC[C@@H](C1)Nc1ncccc1C#N)C(C3)C2 |r,wU:9.10,wD:16.19,3.2,TLB:6:5:29:8.7.9,6:7:4.5.28:29,THB:9:7:4:28.27.29,9:27:4:8.6.7,10:9:4.5.28:29,(2.39,-49.17,;3.73,-48.39,;3.74,-46.85,;5.07,-49.17,;3.88,-50.45,;5.38,-50.03,;6.78,-50.59,;7.8,-49.31,;6.4,-49.66,;7.81,-47.79,;9.15,-47.03,;10.47,-47.81,;10.46,-49.35,;11.82,-47.05,;11.84,-45.51,;13.31,-45.06,;14.2,-46.32,;13.27,-47.55,;15.74,-46.34,;16.53,-45.02,;15.78,-43.69,;16.56,-42.36,;18.11,-42.39,;18.86,-43.73,;18.07,-45.05,;18.82,-46.4,;19.56,-47.74,;6.41,-47.21,;5.37,-48.44,;5.06,-47.69,)| | ||
| Structure | 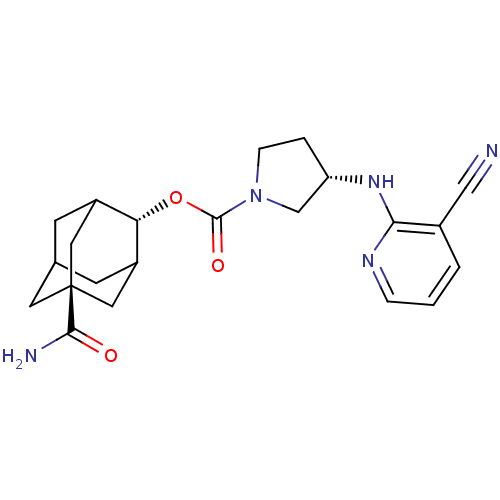
| ||