

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Acetylcholinesterase | ||
| Ligand | BDBM50342868 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_745138 (CHEMBL1772317) | ||
| IC50 | 183±n/a nM | ||
| Citation |  Conejo-García, A; Pisani, L; Núñez, Mdel C; Catto, M; Nicolotti, O; Leonetti, F; Campos, JM; Gallo, MA; Espinosa, A; Carotti, A Homodimeric bis-quaternary heterocyclic ammonium salts as potent acetyl- and butyrylcholinesterase inhibitors: a systematic investigation of the influence of linker and cationic heads over affinity and selectivity. J Med Chem54:2627-45 (2011) [PubMed] Article Conejo-García, A; Pisani, L; Núñez, Mdel C; Catto, M; Nicolotti, O; Leonetti, F; Campos, JM; Gallo, MA; Espinosa, A; Carotti, A Homodimeric bis-quaternary heterocyclic ammonium salts as potent acetyl- and butyrylcholinesterase inhibitors: a systematic investigation of the influence of linker and cationic heads over affinity and selectivity. J Med Chem54:2627-45 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Acetylcholinesterase | |||
| Name: | Acetylcholinesterase | ||
| Synonyms: | ACES_BOVIN | ACHE | Acetylcholinesterase (AChE) | Acetylcholinesterase precursor | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 67659.62 | ||
| Organism: | Bos taurus (bovine) | ||
| Description: | n/a | ||
| Residue: | 613 | ||
| Sequence: |
| ||
| BDBM50342868 | |||
| n/a | |||
| Name | BDBM50342868 | ||
| Synonyms: | 1,1'-(4,4'-methylenebis(4,1-phenylene)bis(methylene))bis(4-(pyrrolidin-1-yl)pyridinium)bromide | CHEMBL1771539 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C33H38N4 | ||
| Mol. Mass. | 490.6805 | ||
| SMILES | C(c1ccc(Cn2ccc(cc2)=[N+]2CCCC2)cc1)c1ccc(Cn2ccc(cc2)=[N+]2CCCC2)cc1 |(1.95,-26.7,;.62,-27.48,;.62,-29.03,;-.72,-29.8,;-2.05,-29.03,;-3.38,-29.8,;-4.72,-29.03,;-4.71,-27.49,;-6.04,-26.72,;-7.38,-27.49,;-7.37,-29.04,;-6.04,-29.8,;-8.71,-26.72,;-8.87,-25.18,;-10.38,-24.86,;-11.15,-26.19,;-10.12,-27.34,;-2.05,-27.49,;-.72,-26.72,;3.28,-27.47,;3.28,-29.01,;4.62,-29.77,;5.95,-29,;7.29,-29.76,;8.62,-28.98,;9.95,-29.75,;11.28,-28.97,;11.28,-27.43,;9.93,-26.67,;8.6,-27.45,;12.6,-26.65,;14.01,-27.26,;15.04,-26.11,;14.26,-24.78,;12.75,-25.11,;5.94,-27.45,;4.6,-26.69,)| | ||
| Structure | 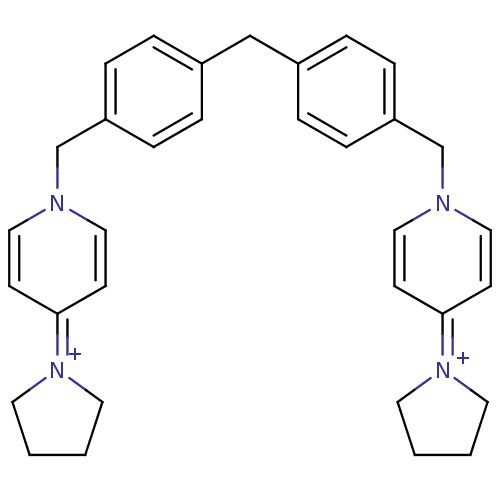
| ||