| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Integrase |
|---|
| Ligand | BDBM50056933 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_90891 (CHEMBL701791) |
|---|
| IC50 | 4900±n/a nM |
|---|
| Citation |  Neamati, N; Hong, H; Mazumder, A; Wang, S; Sunder, S; Nicklaus, MC; Milne, GW; Proksa, B; Pommier, Y Depsides and depsidones as inhibitors of HIV-1 integrase: discovery of novel inhibitors through 3D database searching. J Med Chem40:942-51 (1997) [PubMed] Article Neamati, N; Hong, H; Mazumder, A; Wang, S; Sunder, S; Nicklaus, MC; Milne, GW; Proksa, B; Pommier, Y Depsides and depsidones as inhibitors of HIV-1 integrase: discovery of novel inhibitors through 3D database searching. J Med Chem40:942-51 (1997) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Integrase |
|---|
| Name: | Integrase |
|---|
| Synonyms: | Human immunodeficiency virus type 1 integrase |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 32231.48 |
|---|
| Organism: | Human immunodeficiency virus 1 |
|---|
| Description: | ChEMBL_90865 |
|---|
| Residue: | 288 |
|---|
| Sequence: | FLDGIDKAQDEHEKYHSNWRAMASDFNLPPVVAKEIVASCDKCQLKGEAMHGQVDCSPGI
WQLDCTHLEGKVILVAVHVASGYIEAEVIPAETGQETAYFLLKLAGRWPVKTIHTDNGSN
FTSTTVKAACWWAGIKQEFGIPYNPQSQGVVESMNKELKKIIGQVRDQAEHLKTAVQMAV
FIHNFKRKGGIGGYSAGERIVDIIATDIQTKELQKQITKIQNFRVYYRDSRDPLWKGPAK
LLWKGEGAVVIQDNSDIKVVPRRKVKIIRDYGKQMAGDDCVASRQDED
|
|
|
|---|
| BDBM50056933 |
|---|
| n/a |
|---|
| Name | BDBM50056933 |
|---|
| Synonyms: | (E)-But-2-enedioic acid mono-(7-carboxy-4-formyl-3,8-dihydroxy-1,6-dimethyl-11-oxo-11H-dibenzo[b,e][1,4]dioxepin-9-ylmethyl) ester | CHEMBL366861 | NSC-685588 | fumarprotocetraric acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H16O12 |
|---|
| Mol. Mass. | 472.3552 |
|---|
| SMILES | Cc1cc(O)c(C=O)c2Oc3c(C)c(C(O)=O)c(O)c(COC(=O)\C=C\C(O)=O)c3OC(=O)c12 |
|---|
| Structure | 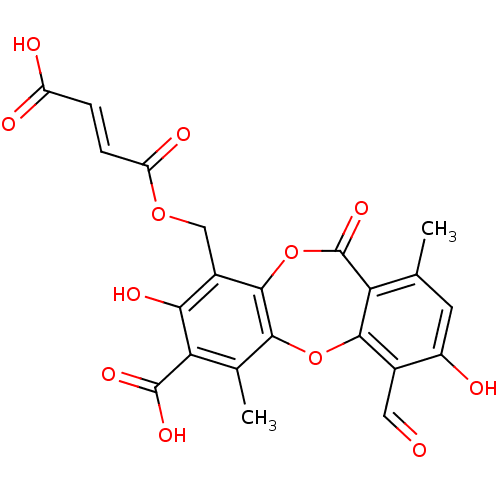
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Neamati, N; Hong, H; Mazumder, A; Wang, S; Sunder, S; Nicklaus, MC; Milne, GW; Proksa, B; Pommier, Y Depsides and depsidones as inhibitors of HIV-1 integrase: discovery of novel inhibitors through 3D database searching. J Med Chem40:942-51 (1997) [PubMed] Article
Neamati, N; Hong, H; Mazumder, A; Wang, S; Sunder, S; Nicklaus, MC; Milne, GW; Proksa, B; Pommier, Y Depsides and depsidones as inhibitors of HIV-1 integrase: discovery of novel inhibitors through 3D database searching. J Med Chem40:942-51 (1997) [PubMed] Article