

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Integrase | ||
| Ligand | BDBM50073642 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_88621 (CHEMBL701725) | ||
| IC50 | 1010±n/a nM | ||
| Citation |  King, PJ; Ma, G; Miao, W; Jia, Q; McDougall, BR; Reinecke, MG; Cornell, C; Kuan, J; Kim, TR; Robinson, WE Structure-activity relationships: analogues of the dicaffeoylquinic and dicaffeoyltartaric acids as potent inhibitors of human immunodeficiency virus type 1 integrase and replication. J Med Chem42:497-509 (1999) [PubMed] Article King, PJ; Ma, G; Miao, W; Jia, Q; McDougall, BR; Reinecke, MG; Cornell, C; Kuan, J; Kim, TR; Robinson, WE Structure-activity relationships: analogues of the dicaffeoylquinic and dicaffeoyltartaric acids as potent inhibitors of human immunodeficiency virus type 1 integrase and replication. J Med Chem42:497-509 (1999) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Integrase | |||
| Name: | Integrase | ||
| Synonyms: | Human immunodeficiency virus type 1 integrase | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 32231.48 | ||
| Organism: | Human immunodeficiency virus 1 | ||
| Description: | ChEMBL_90865 | ||
| Residue: | 288 | ||
| Sequence: |
| ||
| BDBM50073642 | |||
| n/a | |||
| Name | BDBM50073642 | ||
| Synonyms: | (Z)-3-(3,4-Dihydroxy-phenyl)-acrylic acid 4-[(Z)-3-(3,4-dihydroxy-phenyl)-acryloyloxy]-cyclohexyl ester | CHEMBL146489 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C24H24O8 | ||
| Mol. Mass. | 440.4426 | ||
| SMILES | Oc1ccc(\C=C/C(=O)OC2CCC(CC2)OC(=O)\C=C/c2ccc(O)c(O)c2)cc1O |(2.82,-12.69,;2.78,-11.15,;4.09,-10.34,;4.04,-8.83,;2.71,-8.06,;2.69,-6.54,;4.02,-5.76,;5.37,-6.52,;5.37,-8.06,;6.7,-5.75,;8.24,-5.69,;9.04,-7,;10.58,-6.95,;11.32,-5.61,;10.51,-4.3,;8.97,-4.34,;12.86,-5.56,;14.19,-4.79,;14.19,-3.25,;15.52,-5.56,;16.85,-4.79,;16.85,-3.25,;18.2,-2.48,;18.2,-.94,;16.85,-.17,;16.85,1.37,;15.52,-.96,;14.19,-.17,;15.52,-2.48,;1.38,-8.87,;1.42,-10.41,;.1,-11.21,)| | ||
| Structure | 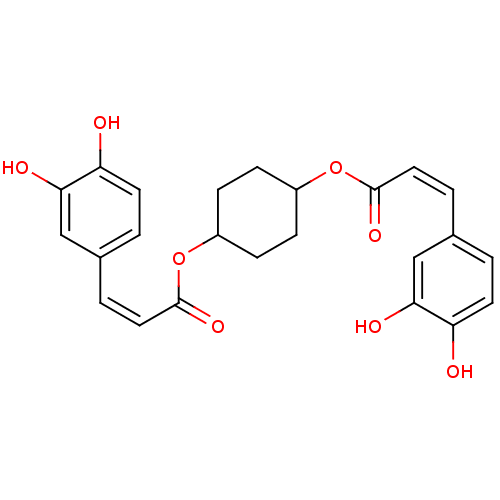
| ||