| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacetylase HST2 |
|---|
| Ligand | BDBM27507 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_606799 (CHEMBL1067344) |
|---|
| IC50 | 91000±n/a nM |
|---|
| Citation |  Sanders, BD; Jackson, B; Brent, M; Taylor, AM; Dang, W; Berger, SL; Schreiber, SL; Howitz, K; Marmorstein, R Identification and characterization of novel sirtuin inhibitor scaffolds. Bioorg Med Chem17:7031-41 (2009) [PubMed] Article Sanders, BD; Jackson, B; Brent, M; Taylor, AM; Dang, W; Berger, SL; Schreiber, SL; Howitz, K; Marmorstein, R Identification and characterization of novel sirtuin inhibitor scaffolds. Bioorg Med Chem17:7031-41 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacetylase HST2 |
|---|
| Name: | NAD-dependent protein deacetylase HST2 |
|---|
| Synonyms: | HST2 | HST2_YEAST | NAD-dependent deacetylase HST2 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 39972.79 |
|---|
| Organism: | Saccharomyces cerevisiae |
|---|
| Description: | ChEMBL_533704 |
|---|
| Residue: | 357 |
|---|
| Sequence: | MSVSTASTEMSVRKIAAHMKSNPNAKVIFMVGAGISTSCGIPDFRSPGTGLYHNLARLKL
PYPEAVFDVDFFQSDPLPFYTLAKELYPGNFRPSKFHYLLKLFQDKDVLKRVYTQNIDTL
ERQAGVKDDLIIEAHGSFAHCHCIGCGKVYPPQVFKSKLAEHPIKDFVKCDVCGELVKPA
IVFFGEDLPDSFSETWLNDSEWLREKITTSGKHPQQPLVIVVGTSLAVYPFASLPEEIPR
KVKRVLCNLETVGDFKANKRPTDLIVHQYSDEFAEQLVEELGWQEDFEKILTAQGGMGDN
SKEQLLEIVHDLENLSLDQSEHESADKKDKKLQRLNGHDSDEDGASNSSSSQKAAKE
|
|
|
|---|
| BDBM27507 |
|---|
| n/a |
|---|
| Name | BDBM27507 |
|---|
| Synonyms: | 3-Pyridinecarboxamide | CHEMBL1140 | NAM | niacinamide | nicotinamide | pyridine-3-carboxamide |
|---|
| Type | n/a |
|---|
| Emp. Form. | C6H6N2O |
|---|
| Mol. Mass. | 122.1246 |
|---|
| SMILES | NC(=O)c1cccnc1 |
|---|
| Structure | 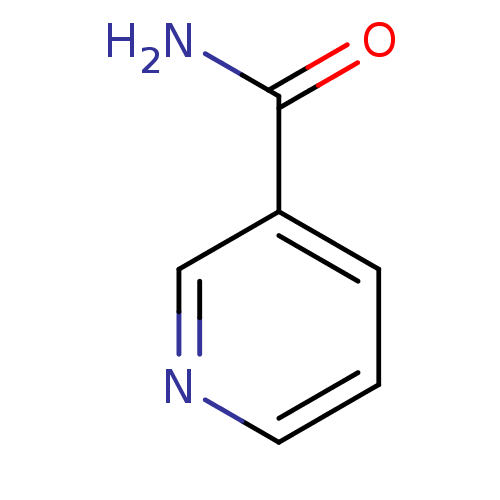
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Sanders, BD; Jackson, B; Brent, M; Taylor, AM; Dang, W; Berger, SL; Schreiber, SL; Howitz, K; Marmorstein, R Identification and characterization of novel sirtuin inhibitor scaffolds. Bioorg Med Chem17:7031-41 (2009) [PubMed] Article
Sanders, BD; Jackson, B; Brent, M; Taylor, AM; Dang, W; Berger, SL; Schreiber, SL; Howitz, K; Marmorstein, R Identification and characterization of novel sirtuin inhibitor scaffolds. Bioorg Med Chem17:7031-41 (2009) [PubMed] Article