| Reaction Details |
|---|
 | Report a problem with these data |
| Target | E3 ubiquitin-protein ligase Mdm2 [1-118] |
|---|
| Ligand | BDBM207980 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | NMR Measurements |
|---|
| pH | 7.4±n/a |
|---|
| Temperature | 288.15±n/a K |
|---|
| Kd | 3.65e+5±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Surmiak, E; Twarda-Clapa, A; Zak, KM; Musielak, B; Tomala, MD; Kubica, K; Grudnik, P; Madej, M; Jablonski, M; Potempa, J; Kalinowska-Tluscik, J; Dömling, A; Dubin, G; Holak, TA A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction. ACS Chem Biol11:3310-3318 (2016) [PubMed] Article Surmiak, E; Twarda-Clapa, A; Zak, KM; Musielak, B; Tomala, MD; Kubica, K; Grudnik, P; Madej, M; Jablonski, M; Potempa, J; Kalinowska-Tluscik, J; Dömling, A; Dubin, G; Holak, TA A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction. ACS Chem Biol11:3310-3318 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| E3 ubiquitin-protein ligase Mdm2 [1-118] |
|---|
| Name: | E3 ubiquitin-protein ligase Mdm2 [1-118] |
|---|
| Synonyms: | MDM2 | MDM2_HUMAN |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 13471.41 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Mdm2 truncation (1-118 aa) |
|---|
| Residue: | 118 |
|---|
| Sequence: | MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQY
IMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDS
|
|
|
|---|
| BDBM207980 |
|---|
| n/a |
|---|
| Name | BDBM207980 |
|---|
| Synonyms: | N-Methyl-5-(4-chlorobenzyl)-4-(6-chloro-1-methyl-1H-indol-3-yl)-3-(4-chlorophenyl)-5-hydroxy-1,5-dihydro-2H-pyrrol-2-one (1e) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H21Cl3N2O2 |
|---|
| Mol. Mass. | 511.827 |
|---|
| SMILES | CN1C(=O)C(=C(c2cn(C)c3cc(Cl)ccc23)C1(O)Cc1ccc(Cl)cc1)c1ccc(Cl)cc1 |t:4| |
|---|
| Structure | 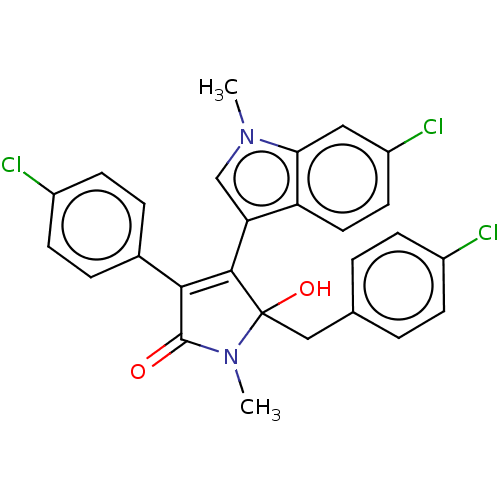
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Surmiak, E; Twarda-Clapa, A; Zak, KM; Musielak, B; Tomala, MD; Kubica, K; Grudnik, P; Madej, M; Jablonski, M; Potempa, J; Kalinowska-Tluscik, J; Dömling, A; Dubin, G; Holak, TA A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction. ACS Chem Biol11:3310-3318 (2016) [PubMed] Article
Surmiak, E; Twarda-Clapa, A; Zak, KM; Musielak, B; Tomala, MD; Kubica, K; Grudnik, P; Madej, M; Jablonski, M; Potempa, J; Kalinowska-Tluscik, J; Dömling, A; Dubin, G; Holak, TA A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction. ACS Chem Biol11:3310-3318 (2016) [PubMed] Article