| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate receptor ionotropic, NMDA 1 |
|---|
| Ligand | BDBM23167 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_140304 |
|---|
| IC50 | 670.0±n/a nM |
|---|
| Citation |  Cordi, A; Lacoste, JM; Audinot, V; Millan, M Design, synthesis and structure-activity relationships of novel strychnine-insensitive glycine receptor ligands. Bioorg Med Chem Lett9:1409-14 (1999) [PubMed] Cordi, A; Lacoste, JM; Audinot, V; Millan, M Design, synthesis and structure-activity relationships of novel strychnine-insensitive glycine receptor ligands. Bioorg Med Chem Lett9:1409-14 (1999) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glutamate receptor ionotropic, NMDA 1 |
|---|
| Name: | Glutamate receptor ionotropic, NMDA 1 |
|---|
| Synonyms: | Glutamate (NMDA) receptor subunit zeta 1 | Glutamate [NMDA] receptor subunit zeta-1 | Glutamate-NMDA-Channel | Glutamate-NMDA-MK801 | Glutamate-NMDA-Polyamine | Grin1 | NMDA | NMDZ1_RAT | Nmdar1 | phencyclidine |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 105533.40 |
|---|
| Organism: | RAT |
|---|
| Description: | P35439 |
|---|
| Residue: | 938 |
|---|
| Sequence: | MSTMHLLTFALLFSCSFARAACDPKIVNIGAVLSTRKHEQMFREAVNQANKRHGSWKIQL
NATSVTHKPNAIQMALSVCEDLISSQVYAILVSHPPTPNDHFTPTPVSYTAGFYRIPVLG
LTTRMSIYSDKSIHLSFLRTVPPYSHQSSVWFEMMRVYNWNHIILLVSDDHEGRAAQKRL
ETLLEERESKAEKVLQFDPGTKNVTALLMEARELEARVIILSASEDDAATVYRAAAMLNM
TGSGYVWLVGEREISGNALRYAPDGIIGLQLINGKNESAHISDAVGVVAQAVHELLEKEN
ITDPPRGCVGNTNIWKTGPLFKRVLMSSKYADGVTGRVEFNEDGDRKFANYSIMNLQNRK
LVQVGIYNGTHVIPNDRKIIWPGGETEKPRGYQMSTRLKIVTIHQEPFVYVKPTMSDGTC
KEEFTVNGDPVKKVICTGPNDTSPGSPRHTVPQCCYGFCIDLLIKLARTMNFTYEVHLVA
DGKFGTQERVNNSNKKEWNGMMGELLSGQADMIVAPLTINNERAQYIEFSKPFKYQGLTI
LVKKEIPRSTLDSFMQPFQSTLWLLVGLSVHVVAVMLYLLDRFSPFGRFKVNSEEEEEDA
LTLSSAMWFSWGVLLNSGIGEGAPRSFSARILGMVWAGFAMIIVASYTANLAAFLVLDRP
EERITGINDPRLRNPSDKFIYATVKQSSVDIYFRRQVELSTMYRHMEKHNYESAAEAIQA
VRDNKLHAFIWDSAVLEFEASQKCDLVTTGELFFRSGFGIGMRKDSPWKQNVSLSILKSH
ENGFMEDLDKTWVRYQECDSRSNAPATLTFENMAGVFMLVAGGIVAGIFLIFIEIAYKRH
KDARRKQMQLAFAAVNVWRKNLQDRKSGRAEPDPKKKATFRAITSTLASSFKRRRSSKDT
STGGGRGALQNQKDTVLPRRAIEREEGQLQLCSRHRES
|
|
|
|---|
| BDBM23167 |
|---|
| n/a |
|---|
| Name | BDBM23167 |
|---|
| Synonyms: | (2R)-2-amino-3-hydroxypropanoic acid | CHEMBL285123 | D-Serine |
|---|
| Type | Amino Acid |
|---|
| Emp. Form. | C3H7NO3 |
|---|
| Mol. Mass. | 105.0926 |
|---|
| SMILES | N[C@H](CO)C(O)=O |
|---|
| Structure | 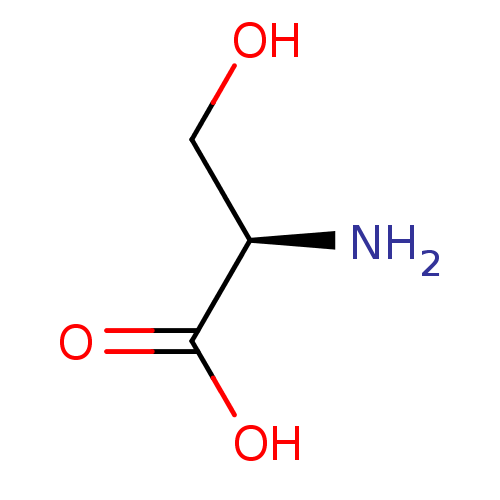
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cordi, A; Lacoste, JM; Audinot, V; Millan, M Design, synthesis and structure-activity relationships of novel strychnine-insensitive glycine receptor ligands. Bioorg Med Chem Lett9:1409-14 (1999) [PubMed]
Cordi, A; Lacoste, JM; Audinot, V; Millan, M Design, synthesis and structure-activity relationships of novel strychnine-insensitive glycine receptor ligands. Bioorg Med Chem Lett9:1409-14 (1999) [PubMed]