| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Sodium- and chloride-dependent glycine transporter 2 |
|---|
| Ligand | BDBM50263396 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_536095 (CHEMBL995041) |
|---|
| EC50 | 4200±n/a nM |
|---|
| Citation |  Jolidon, S; Alberati, D; Dowle, A; Fischer, H; Hainzl, D; Narquizian, R; Norcross, R; Pinard, E Design, synthesis and structure-activity relationship of simple bis-amides as potent inhibitors of GlyT1. Bioorg Med Chem Lett18:5533-6 (2008) [PubMed] Article Jolidon, S; Alberati, D; Dowle, A; Fischer, H; Hainzl, D; Narquizian, R; Norcross, R; Pinard, E Design, synthesis and structure-activity relationship of simple bis-amides as potent inhibitors of GlyT1. Bioorg Med Chem Lett18:5533-6 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Sodium- and chloride-dependent glycine transporter 2 |
|---|
| Name: | Sodium- and chloride-dependent glycine transporter 2 |
|---|
| Synonyms: | GLYT2 | Glycine transporter 2 | NET1 | SC6A5_HUMAN | SLC6A5 | Sodium- and chloride-dependent glycine transporter 2 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 87438.56 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Glycine-2-transporter 0 HUMAN::Q9Y345 |
|---|
| Residue: | 797 |
|---|
| Sequence: | MDCSAPKEMNKLPANSPEAAAAQGHPDGPCAPRTSPEQELPAAAAPPPPRVPRSASTGAQ
TFQSADARACEAERPGVGSCKLSSPRAQAASAALRDLREAQGAQASPPPGSSGPGNALHC
KIPFLRGPEGDANVSVGKGTLERNNTPVVGWVNMSQSTVVLATDGITSVLPGSVATVATQ
EDEQGDENKARGNWSSKLDFILSMVGYAVGLGNVWRFPYLAFQNGGGAFLIPYLMMLALA
GLPIFFLEVSLGQFASQGPVSVWKAIPALQGCGIAMLIISVLIAIYYNVIICYTLFYLFA
SFVSVLPWGSCNNPWNTPECKDKTKLLLDSCVISDHPKIQIKNSTFCMTAYPNVTMVNFT
SQANKTFVSGSEEYFKYFVLKISAGIEYPGEIRWPLALCLFLAWVIVYASLAKGIKTSGK
VVYFTATFPYVVLVILLIRGVTLPGAGAGIWYFITPKWEKLTDATVWKDAATQIFFSLSA
AWGGLITLSSYNKFHNNCYRDTLIVTCTNSATSIFAGFVIFSVIGFMANERKVNIENVAD
QGPGIAFVVYPEALTRLPLSPFWAIIFFLMLLTLGLDTMFATIETIVTSISDEFPKYLRT
HKPVFTLGCCICFFIMGFPMITQGGIYMFQLVDTYAASYALVIIAIFELVGISYVYGLQR
FCEDIEMMIGFQPNIFWKVCWAFVTPTILTFILCFSFYQWEPMTYGSYRYPNWSMVLGWL
MLACSVIWIPIMFVIKMHLAPGRFIERLKLVCSPQPDWGPFLAQHRGERYKNMIDPLGTS
SLGLKLPVKDLELGTQC
|
|
|
|---|
| BDBM50263396 |
|---|
| n/a |
|---|
| Name | BDBM50263396 |
|---|
| Synonyms: | CHEMBL476763 | N-(2-((4-chlorophenyl)(phenyl)methylamino)-2-oxoethyl)benzamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H19ClN2O2 |
|---|
| Mol. Mass. | 378.851 |
|---|
| SMILES | Clc1ccc(cc1)C(NC(=O)CNC(=O)c1ccccc1)c1ccccc1 |
|---|
| Structure | 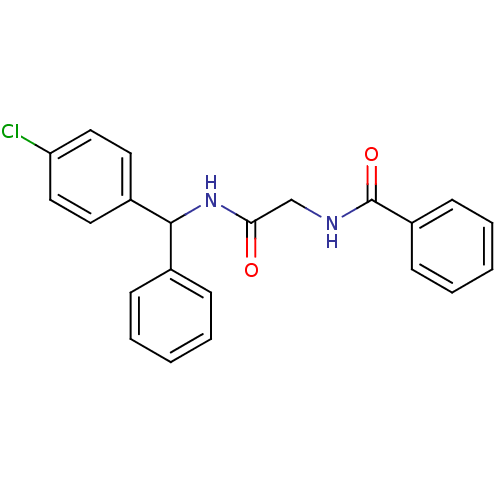
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jolidon, S; Alberati, D; Dowle, A; Fischer, H; Hainzl, D; Narquizian, R; Norcross, R; Pinard, E Design, synthesis and structure-activity relationship of simple bis-amides as potent inhibitors of GlyT1. Bioorg Med Chem Lett18:5533-6 (2008) [PubMed] Article
Jolidon, S; Alberati, D; Dowle, A; Fischer, H; Hainzl, D; Narquizian, R; Norcross, R; Pinard, E Design, synthesis and structure-activity relationship of simple bis-amides as potent inhibitors of GlyT1. Bioorg Med Chem Lett18:5533-6 (2008) [PubMed] Article