

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 2C19 | ||
| Ligand | BDBM50419619 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_797372 (CHEMBL1942948) | ||
| IC50 | <10000±n/a nM | ||
| Citation |  De Savi, C; Morley, AD; Nash, I; Karoutchi, G; Page, K; Ting, A; Gerhardt, S Lead optimisation of selective non-zinc binding inhibitors of MMP13. Part 2. Bioorg Med Chem Lett22:271-7 (2011) [PubMed] Article De Savi, C; Morley, AD; Nash, I; Karoutchi, G; Page, K; Ting, A; Gerhardt, S Lead optimisation of selective non-zinc binding inhibitors of MMP13. Part 2. Bioorg Med Chem Lett22:271-7 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 2C19 | |||
| Name: | Cytochrome P450 2C19 | ||
| Synonyms: | (R)-limonene 6-monooxygenase | (S)-limonene 6-monooxygenase | (S)-limonene 7-monooxygenase | CP2CJ_HUMAN | CYP2C19 | CYPIIC17 | CYPIIC19 | Cytochrome P450 2C19 (CYP2C19) | Cytochrome P450 2C19 [I331V] | Cytochrome P450-11A | Cytochrome P450-254C | Fenbendazole monooxygenase (4'-hydroxylating) | Mephenytoin 4-hydroxylase | P450-11A | P450-254C | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 55935.47 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P33261 | ||
| Residue: | 490 | ||
| Sequence: |
| ||
| BDBM50419619 | |||
| n/a | |||
| Name | BDBM50419619 | ||
| Synonyms: | CHEMBL1940296 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H24N2O3 | ||
| Mol. Mass. | 376.4483 | ||
| SMILES | CNC(=O)c1ccc(Oc2ccc(cc2)C#C[C@]2(O)CN3CCC2CC3)cc1 |r,wU:17.18,wD:17.17,(18.36,-56.75,;19.7,-57.53,;21.04,-56.76,;21.04,-55.22,;22.37,-57.54,;23.71,-56.78,;25.04,-57.55,;25.03,-59.09,;26.36,-59.87,;27.7,-59.11,;27.71,-57.56,;29.05,-56.8,;30.38,-57.59,;30.37,-59.12,;29.04,-59.88,;31.72,-56.83,;33.06,-56.07,;34.37,-55.28,;34.35,-56.8,;34.34,-53.75,;35.66,-52.97,;37.78,-53.4,;37.82,-55.58,;35.69,-56.02,;36.98,-55.24,;36.97,-53.74,;23.7,-59.86,;22.37,-59.09,)| | ||
| Structure | 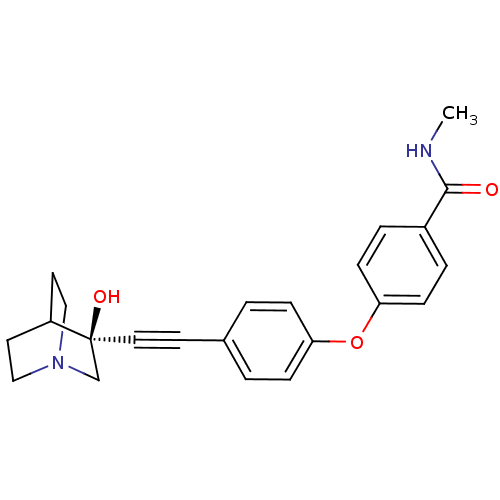
| ||