| Reaction Details |
|---|
 | Report a problem with these data |
| Target | High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A |
|---|
| Ligand | BDBM99145 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1437290 (CHEMBL3384793) |
|---|
| IC50 | >30000±n/a nM |
|---|
| Citation |  Rzasa, RM; Frohn, MJ; Andrews, KL; Chmait, S; Chen, N; Clarine, JG; Davis, C; Eastwood, HA; Horne, DB; Hu, E; Jones, AD; Kaller, MR; Kunz, RK; Miller, S; Monenschein, H; Nguyen, T; Pickrell, AJ; Porter, A; Reichelt, A; Zhao, X; Treanor, JJ; Allen, JR Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility. Bioorg Med Chem22:6570-85 (2015) [PubMed] Article Rzasa, RM; Frohn, MJ; Andrews, KL; Chmait, S; Chen, N; Clarine, JG; Davis, C; Eastwood, HA; Horne, DB; Hu, E; Jones, AD; Kaller, MR; Kunz, RK; Miller, S; Monenschein, H; Nguyen, T; Pickrell, AJ; Porter, A; Reichelt, A; Zhao, X; Treanor, JJ; Allen, JR Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility. Bioorg Med Chem22:6570-85 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A |
|---|
| Name: | High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A |
|---|
| Synonyms: | High-affinity cAMP-specific and IBMX-insensitive 3 ,5 -cyclic phosphodiesterase 8A | PDE8A | PDE8A_HUMAN | Phosphodiesterase 8 | Phosphodiesterase 8 (PDE8) | Phosphodiesterase 8A (PDE8A) | Phosphodiesterase 8A (PDE8A1) | Phosphodiesterase 8A1 | Phosphodiesterase Type 8 (PDE8A) |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 93295.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Recombinant catalytic domain (M1-E829) of human PDE8A. |
|---|
| Residue: | 829 |
|---|
| Sequence: | MGCAPSIHISERLVAEDAPSPAAPPLSSGGPRLPQGQKTAALPRTRGAGLLESELRDGSG
KKVAVADVQFGPMRFHQDQLQVLLVFTKEDNQCNGFCRACEKAGFKCTVTKEAQAVLACF
LDKHHDIIIIDHRNPRQLDAEALCRSIRSSKLSENTVIVGVVRRVDREELSVMPFISAGF
TRRYVENPNIMACYNELLQLEFGEVRSQLKLRACNSVFTALENSEDAIEITSEDRFIQYA
NPAFETTMGYQSGELIGKELGEVPINEKKADLLDTINSCIRIGKEWQGIYYAKKKNGDNI
QQNVKIIPVIGQGGKIRHYVSIIRVCNGNNKAEKISECVQSDTHTDNQTGKHKDRRKGSL
DVKAVASRATEVSSQRRHSSMARIHSMTIEAPITKVINIINAAQESSPMPVTEALDRVLE
ILRTTELYSPQFGAKDDDPHANDLVGGLMSDGLRRLSGNEYVLSTKNTQMVSSNIITPIS
LDDVPPRIARAMENEEYWDFDIFELEAATHNRPLIYLGLKMFARFGICEFLHCSESTLRS
WLQIIEANYHSSNPYHNSTHSADVLHATAYFLSKERIKETLDPIDEVAALIAATIHDVDH
PGRTNSFLCNAGSELAILYNDTAVLESHHAALAFQLTTGDDKCNIFKNMERNDYRTLRQG
IIDMVLATEMTKHFEHVNKFVNSINKPLATLEENGETDKNQEVINTMLRTPENRTLIKRM
LIKCADVSNPCRPLQYCIEWAARISEEYFSQTDEEKQQGLPVVMPVFDRNTCSIPKSQIS
FIDYFITDMFDAWDAFVDLPDLMQHLDNNFKYWKGLDEMKLRNLRPPPE
|
|
|
|---|
| BDBM99145 |
|---|
| n/a |
|---|
| Name | BDBM99145 |
|---|
| Synonyms: | US8497265, 894 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H25N5O2 |
|---|
| Mol. Mass. | 403.4769 |
|---|
| SMILES | CC(=O)N1CCC(CC1)c1nccnc1OC1CN(C1)c1ccc2ccccc2n1 |
|---|
| Structure | 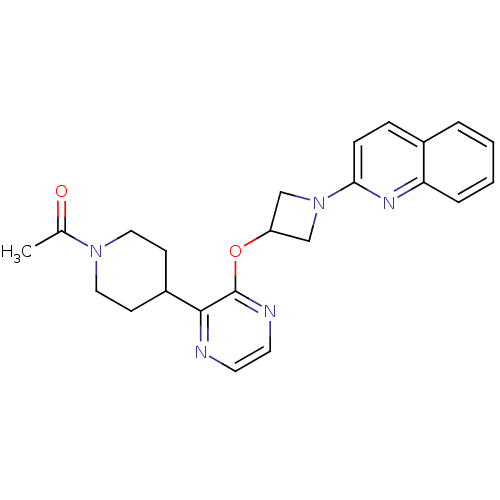
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rzasa, RM; Frohn, MJ; Andrews, KL; Chmait, S; Chen, N; Clarine, JG; Davis, C; Eastwood, HA; Horne, DB; Hu, E; Jones, AD; Kaller, MR; Kunz, RK; Miller, S; Monenschein, H; Nguyen, T; Pickrell, AJ; Porter, A; Reichelt, A; Zhao, X; Treanor, JJ; Allen, JR Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility. Bioorg Med Chem22:6570-85 (2015) [PubMed] Article
Rzasa, RM; Frohn, MJ; Andrews, KL; Chmait, S; Chen, N; Clarine, JG; Davis, C; Eastwood, HA; Horne, DB; Hu, E; Jones, AD; Kaller, MR; Kunz, RK; Miller, S; Monenschein, H; Nguyen, T; Pickrell, AJ; Porter, A; Reichelt, A; Zhao, X; Treanor, JJ; Allen, JR Synthesis and preliminary biological evaluation of potent and selective 2-(3-alkoxy-1-azetidinyl) quinolines as novel PDE10A inhibitors with improved solubility. Bioorg Med Chem22:6570-85 (2015) [PubMed] Article