| Citation |  Madar, DJ; Kopecka, H; Pireh, D; Yong, H; Pei, Z; Li, X; Wiedeman, PE; Djuric, SW; Von Geldern, TW; Fickes, MG; Bhagavatula, L; McDermott, T; Wittenberger, S; Richards, SJ; Longenecker, KL; Stewart, KD; Lubben, TH; Ballaron, SJ; Stashko, MA; Long, MA; Wells, H; Zinker, BA; Mika, AK; Beno, DW; Kempf-Grote, AJ; Polakowski, J; Segreti, J; Reinhart, GA; Fryer, RM; Sham, HL; Trevillyan, JM Discovery of 2-[4-{{2-(2S,5R)-2-cyano-5-ethynyl-1-pyrrolidinyl]-2-oxoethyl]amino]-4-methyl-1-piperidinyl]-4-pyridinecarboxylic acid (ABT-279): a very potent, selective, effective, and well-tolerated inhibitor of dipeptidyl peptidase-IV, useful for the treatment of diabetes. J Med Chem49:6416-20 (2006) [PubMed] Article Madar, DJ; Kopecka, H; Pireh, D; Yong, H; Pei, Z; Li, X; Wiedeman, PE; Djuric, SW; Von Geldern, TW; Fickes, MG; Bhagavatula, L; McDermott, T; Wittenberger, S; Richards, SJ; Longenecker, KL; Stewart, KD; Lubben, TH; Ballaron, SJ; Stashko, MA; Long, MA; Wells, H; Zinker, BA; Mika, AK; Beno, DW; Kempf-Grote, AJ; Polakowski, J; Segreti, J; Reinhart, GA; Fryer, RM; Sham, HL; Trevillyan, JM Discovery of 2-[4-{{2-(2S,5R)-2-cyano-5-ethynyl-1-pyrrolidinyl]-2-oxoethyl]amino]-4-methyl-1-piperidinyl]-4-pyridinecarboxylic acid (ABT-279): a very potent, selective, effective, and well-tolerated inhibitor of dipeptidyl peptidase-IV, useful for the treatment of diabetes. J Med Chem49:6416-20 (2006) [PubMed] Article |
|---|
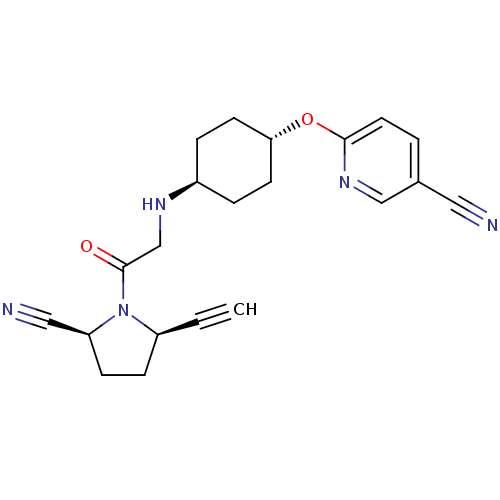
| SMILES | O=C(CN[C@H]1CC[C@@H](CC1)Oc1ccc(cn1)C#N)N1[C@H](CC[C@H]1C#N)C#C |r,wU:23.26,20.28,7.10,wD:4.3,(-4.29,1.85,;-4.29,3.39,;-5.62,4.16,;-6.95,3.39,;-8.29,4.16,;-8.25,5.7,;-9.56,6.51,;-10.92,5.77,;-10.96,4.23,;-9.64,3.43,;-12.23,6.57,;-13.72,6.17,;-14.35,4.77,;-15.88,4.61,;-16.78,5.86,;-16.15,7.26,;-14.62,7.42,;-18.27,5.46,;-19.76,5.06,;-2.95,4.16,;-2.26,5.54,;-.74,5.3,;-.49,3.78,;-1.86,3.08,;-2.26,1.59,;-2.66,.1,;-3.03,6.87,;-3.8,8.2,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Madar, DJ; Kopecka, H; Pireh, D; Yong, H; Pei, Z; Li, X; Wiedeman, PE; Djuric, SW; Von Geldern, TW; Fickes, MG; Bhagavatula, L; McDermott, T; Wittenberger, S; Richards, SJ; Longenecker, KL; Stewart, KD; Lubben, TH; Ballaron, SJ; Stashko, MA; Long, MA; Wells, H; Zinker, BA; Mika, AK; Beno, DW; Kempf-Grote, AJ; Polakowski, J; Segreti, J; Reinhart, GA; Fryer, RM; Sham, HL; Trevillyan, JM Discovery of 2-[4-{{2-(2S,5R)-2-cyano-5-ethynyl-1-pyrrolidinyl]-2-oxoethyl]amino]-4-methyl-1-piperidinyl]-4-pyridinecarboxylic acid (ABT-279): a very potent, selective, effective, and well-tolerated inhibitor of dipeptidyl peptidase-IV, useful for the treatment of diabetes. J Med Chem49:6416-20 (2006) [PubMed] Article
Madar, DJ; Kopecka, H; Pireh, D; Yong, H; Pei, Z; Li, X; Wiedeman, PE; Djuric, SW; Von Geldern, TW; Fickes, MG; Bhagavatula, L; McDermott, T; Wittenberger, S; Richards, SJ; Longenecker, KL; Stewart, KD; Lubben, TH; Ballaron, SJ; Stashko, MA; Long, MA; Wells, H; Zinker, BA; Mika, AK; Beno, DW; Kempf-Grote, AJ; Polakowski, J; Segreti, J; Reinhart, GA; Fryer, RM; Sham, HL; Trevillyan, JM Discovery of 2-[4-{{2-(2S,5R)-2-cyano-5-ethynyl-1-pyrrolidinyl]-2-oxoethyl]amino]-4-methyl-1-piperidinyl]-4-pyridinecarboxylic acid (ABT-279): a very potent, selective, effective, and well-tolerated inhibitor of dipeptidyl peptidase-IV, useful for the treatment of diabetes. J Med Chem49:6416-20 (2006) [PubMed] Article