| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Methionine aminopeptidase |
|---|
| Ligand | BDBM17868 |
|---|
| Substrate/Competitor | BDBM17848 |
|---|
| Meas. Tech. | MetAP Inhibition Assay |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| IC50 | 16000±1800 nM |
|---|
| Citation |  Douangamath, A; Dale, GE; D'Arcy, A; Almstetter, M; Eckl, R; Frutos-Hoener, A; Henkel, B; Illgen, K; Nerdinger, S; Schulz, H; Mac Sweeney, A; Thormann, M; Treml, A; Pierau, S; Wadman, S; Oefner, C Crystal structures of Staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate. J Med Chem47:1325-8 (2004) [PubMed] Article Douangamath, A; Dale, GE; D'Arcy, A; Almstetter, M; Eckl, R; Frutos-Hoener, A; Henkel, B; Illgen, K; Nerdinger, S; Schulz, H; Mac Sweeney, A; Thormann, M; Treml, A; Pierau, S; Wadman, S; Oefner, C Crystal structures of Staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate. J Med Chem47:1325-8 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Methionine aminopeptidase |
|---|
| Name: | Methionine aminopeptidase |
|---|
| Synonyms: | MAP1_STAAW | MetAP | Methionine Aminopeptidase (MAP) | Peptidase M | SaMetAP-1 | map |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 27494.40 |
|---|
| Organism: | Staphylococcus aureus |
|---|
| Description: | P0A079 |
|---|
| Residue: | 252 |
|---|
| Sequence: | MIVKTEEELQALKEIGYICAKVRNTMQAATKPGITTKELDNIAKELFEEYGAISAPIHDE
NFPGQTCISVNEEVAHGIPSKRVIREGDLVNIDVSALKNGYYADTGISFVVGESDDPMKQ
KVCDVATMAFENAIAKVKPGTKLSNIGKAVHNTARQNDLKVIKNLTGHGVGLSLHEAPAH
VLNYFDPKDKTLLTEGMVLAIEPFISSNASFVTEGKNEWAFETSDKSFVAQIEHTVIVTK
DGPILTTKIEEE
|
|
|
|---|
| BDBM17868 |
|---|
| BDBM17848 |
|---|
| Name | BDBM17868 |
|---|
| Synonyms: | (2S)-2-amino-4-(methylsulfanyl)-1-(pyridin-2-yl)butan-1-one | keto-heterocycle inhibitor, 2 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H14N2OS |
|---|
| Mol. Mass. | 210.296 |
|---|
| SMILES | CSCC[C@H](N)C(=O)c1ccccn1 |r| |
|---|
| Structure | 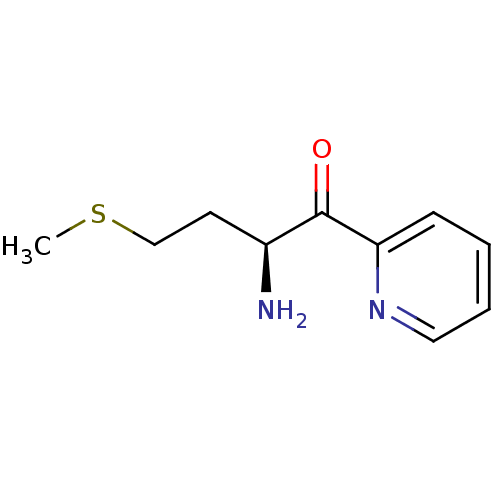
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Douangamath, A; Dale, GE; D'Arcy, A; Almstetter, M; Eckl, R; Frutos-Hoener, A; Henkel, B; Illgen, K; Nerdinger, S; Schulz, H; Mac Sweeney, A; Thormann, M; Treml, A; Pierau, S; Wadman, S; Oefner, C Crystal structures of Staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate. J Med Chem47:1325-8 (2004) [PubMed] Article
Douangamath, A; Dale, GE; D'Arcy, A; Almstetter, M; Eckl, R; Frutos-Hoener, A; Henkel, B; Illgen, K; Nerdinger, S; Schulz, H; Mac Sweeney, A; Thormann, M; Treml, A; Pierau, S; Wadman, S; Oefner, C Crystal structures of Staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate. J Med Chem47:1325-8 (2004) [PubMed] Article