| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phosphoribosylglycinamide formyltransferase |
|---|
| Ligand | BDBM22587 |
|---|
| Substrate/Competitor | BDBM22589 |
|---|
| Meas. Tech. | GAR Tfase Activity Assay |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 299.15±n/a K |
|---|
| Ki | >100000±n/a nM |
|---|
| Citation |  Xu, L; Chong, Y; Hwang, I; D'Onofrio, A; Amore, K; Beardsley, GP; Li, C; Olson, AJ; Boger, DL; Wilson, IA Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase. J Biol Chem282:13033-46 (2007) [PubMed] Article Xu, L; Chong, Y; Hwang, I; D'Onofrio, A; Amore, K; Beardsley, GP; Li, C; Olson, AJ; Boger, DL; Wilson, IA Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase. J Biol Chem282:13033-46 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Phosphoribosylglycinamide formyltransferase |
|---|
| Name: | Phosphoribosylglycinamide formyltransferase |
|---|
| Synonyms: | 5 -phosphoribosylglycinamide transformylase | GAR Tfase | GART | Glycinamide Ribonucleotide Transformylase | PUR3_ECOLI | Phosphoribosylglycinamide formyltransferase | purN |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 23233.97 |
|---|
| Organism: | Escherichia coli (strain K12) |
|---|
| Description: | n/a |
|---|
| Residue: | 212 |
|---|
| Sequence: | MNIVVLISGNGSNLQAIIDACKTNKIKGTVRAVFSNKADAFGLERARQAGIATHTLIASA
FDSREAYDRELIHEIDMYAPDVVVLAGFMRILSPAFVSHYAGRLLNIHPSLLPKYPGLHT
HRQALENGDEEHGTSVHFVTDELDGGPVILQAKVPVFAGDSEDDITARVQTQEHAIYPLV
ISWFADGRLKMHENAAWLDGQRLPPQGYAADE
|
|
|
|---|
| BDBM22587 |
|---|
| BDBM22589 |
|---|
| Name | BDBM22587 |
|---|
| Synonyms: | 7-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1H,3H,4H,7H-2,1,3,5,7-imidazo[4,5-c][1,2,6]thiadiazine-2,2,4-trione | Nucleoside, 2 |
|---|
| Type | Nucleoside or nucleotide |
|---|
| Emp. Form. | C9H12N4O7S |
|---|
| Mol. Mass. | 320.279 |
|---|
| SMILES | OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)n1cnc2c1NS(=O)(=O)NC2=O |r| |
|---|
| Structure | 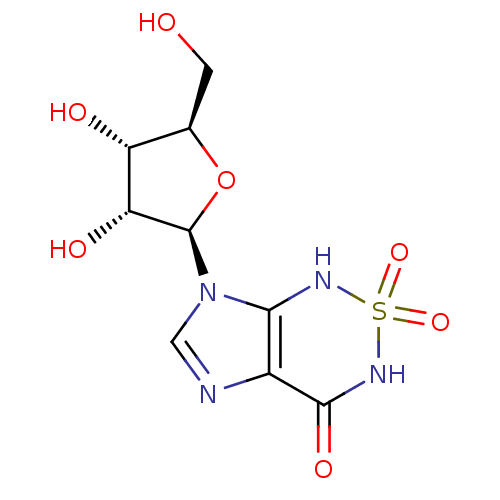
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Xu, L; Chong, Y; Hwang, I; D'Onofrio, A; Amore, K; Beardsley, GP; Li, C; Olson, AJ; Boger, DL; Wilson, IA Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase. J Biol Chem282:13033-46 (2007) [PubMed] Article
Xu, L; Chong, Y; Hwang, I; D'Onofrio, A; Amore, K; Beardsley, GP; Li, C; Olson, AJ; Boger, DL; Wilson, IA Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase. J Biol Chem282:13033-46 (2007) [PubMed] Article