| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Alpha-1B adrenergic receptor |
|---|
| Ligand | BDBM29560 |
|---|
| Substrate/Competitor | BDBM29568 |
|---|
| Meas. Tech. | Radioligand Binding Assay (Ki) |
|---|
| Ki | 101±n/a nM |
|---|
| Citation |  Paluchowska, MH; Bugno, R; Duszyńska, B; Tatarczyńska, E; Nikiforuk, A; Lenda, T; Chojnacka-Wójcik, E The influence of modifications in imide fragment structure on 5-HT(1A) and 5-HT(7) receptor affinity and in vivo pharmacological properties of some new 1-(m-trifluoromethylphenyl)piperazines. Bioorg Med Chem15:7116-25 (2007) [PubMed] Article Paluchowska, MH; Bugno, R; Duszyńska, B; Tatarczyńska, E; Nikiforuk, A; Lenda, T; Chojnacka-Wójcik, E The influence of modifications in imide fragment structure on 5-HT(1A) and 5-HT(7) receptor affinity and in vivo pharmacological properties of some new 1-(m-trifluoromethylphenyl)piperazines. Bioorg Med Chem15:7116-25 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Alpha-1B adrenergic receptor |
|---|
| Name: | Alpha-1B adrenergic receptor |
|---|
| Synonyms: | ADA1B_RAT | Adra1b | Alpha 1B-adrenoceptor | Alpha 1B-adrenoreceptor | Alpha adrenergic receptor 1A and 1B | Alpha-1 Adrenergic Receptor | Alpha-1Adrenoceptor | Alpha-1B adrenergic receptor | Alpha-1B adrenoreceptor | adrenergic Alpha1B |
|---|
| Type: | G Protein-Coupled Receptor (GPCR) |
|---|
| Mol. Mass.: | 56606.71 |
|---|
| Organism: | Rattus norvegicus (rat) |
|---|
| Description: | Receptor binding assays were performed using rat cortical membranes. |
|---|
| Residue: | 515 |
|---|
| Sequence: | MNPDLDTGHNTSAPAHWGELKDDNFTGPNQTSSNSTLPQLDVTRAISVGLVLGAFILFAI
VGNILVILSVACNRHLRTPTNYFIVNLAIADLLLSFTVLPFSATLEVLGYWVLGRIFCDI
WAAVDVLCCTASILSLCAISIDRYIGVRYSLQYPTLVTRRKAILALLSVWVLSTVISIGP
LLGWKEPAPNDDKECGVTEEPFYALFSSLGSFYIPLAVILVMYCRVYIVAKRTTKNLEAG
VMKEMSNSKELTLRIHSKNFHEDTLSSTKAKGHNPRSSIAVKLFKFSREKKAAKTLGIVV
GMFILCWLPFFIALPLGSLFSTLKPPDAVFKVVFWLGYFNSCLNPIIYPCSSKEFKRAFM
RILGCQCRGGRRRRRRRRLGACAYTYRPWTRGGSLERSQSRKDSLDDSGSCMSGTQRTLP
SASPSPGYLGRGTQPPVELCAFPEWKPGALLSLPEPPGRRGRLDSGPLFTFKLLGDPESP
GTEGDTSNGGCDTTTDLANGQPGFKSNMPLAPGHF
|
|
|
|---|
| BDBM29560 |
|---|
| BDBM29568 |
|---|
| Name | BDBM29560 |
|---|
| Synonyms: | 1-(m-trifluorophenyl)piperazine, 7 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H26F3N3O2 |
|---|
| Mol. Mass. | 397.4345 |
|---|
| SMILES | FC(F)(F)c1cccc(c1)N1CCN(CCCCN2C(=O)CCCC2=O)CC1 |
|---|
| Structure | 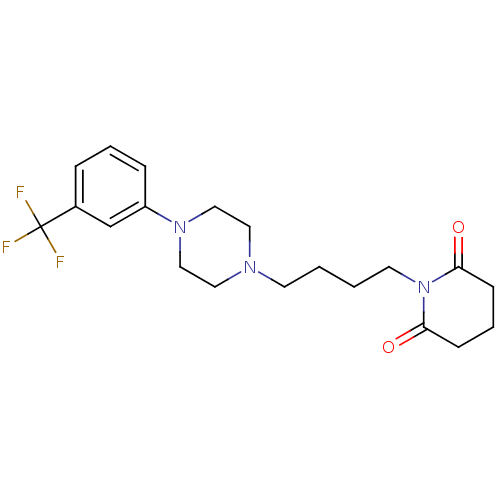
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Paluchowska, MH; Bugno, R; Duszyńska, B; Tatarczyńska, E; Nikiforuk, A; Lenda, T; Chojnacka-Wójcik, E The influence of modifications in imide fragment structure on 5-HT(1A) and 5-HT(7) receptor affinity and in vivo pharmacological properties of some new 1-(m-trifluoromethylphenyl)piperazines. Bioorg Med Chem15:7116-25 (2007) [PubMed] Article
Paluchowska, MH; Bugno, R; Duszyńska, B; Tatarczyńska, E; Nikiforuk, A; Lenda, T; Chojnacka-Wójcik, E The influence of modifications in imide fragment structure on 5-HT(1A) and 5-HT(7) receptor affinity and in vivo pharmacological properties of some new 1-(m-trifluoromethylphenyl)piperazines. Bioorg Med Chem15:7116-25 (2007) [PubMed] Article