| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
|---|
| Ligand | BDBM59101 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Steady-State Inhibition Assay |
|---|
| pH | 7.4±0 |
|---|
| Ki | 7.9e+3±n/a nM |
|---|
| Citation |  Sem, DS; Bertolaet, B; Baker, B; Chang, E; Costache, AD; Coutts, S; Dong, Q; Hansen, M; Hong, V; Huang, X; Jack, RM; Kho, R; Lang, H; Ma, CT; Meininger, D; Pellecchia, M; Pierre, F; Villar, H; Yu, L Systems-based design of bi-ligand inhibitors of oxidoreductases: filling the chemical proteomic toolbox. Chem Biol11:185-94 (2004) [PubMed] Article Sem, DS; Bertolaet, B; Baker, B; Chang, E; Costache, AD; Coutts, S; Dong, Q; Hansen, M; Hong, V; Huang, X; Jack, RM; Kho, R; Lang, H; Ma, CT; Meininger, D; Pellecchia, M; Pierre, F; Villar, H; Yu, L Systems-based design of bi-ligand inhibitors of oxidoreductases: filling the chemical proteomic toolbox. Chem Biol11:185-94 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
|---|
| Name: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
|---|
| Synonyms: | 1-Deoxy-D-xylulose 5-phosphate reductoisomerase (DXP) | 1-Deoxyxylulose 5-phosphate reductoisomerase (DXR) | 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) | 2-C-methyl-D-erythritol 4-phosphate synthase | DXP reductoisomerase | DXR_ECOLI | dxr | ispC | yaeM |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 43384.52 |
|---|
| Organism: | Escherichia coli |
|---|
| Description: | P45568 |
|---|
| Residue: | 398 |
|---|
| Sequence: | MKQLTILGSTGSIGCSTLDVVRHNPEHFRVVALVAGKNVTRMVEQCLEFSPRYAVMDDEA
SAKLLKTMLQQQGSRTEVLSGQQAACDMAALEDVDQVMAAIVGAAGLLPTLAAIRAGKTI
LLANKESLVTCGRLFMDAVKQSKAQLLPVDSEHNAIFQSLPQPIQHNLGYADLEQNGVVS
ILLTGSGGPFRETPLRDLATMTPDQACRHPNWSMGRKISVDSATMMNKGLEYIEARWLFN
ASASQMEVLIHPQSVIHSMVRYQDGSVLAQLGEPDMRTPIAHTMAWPNRVNSGVKPLDFC
KLSALTFAAPDYDRYPCLKLAMEAFEQGQAATTALNAANEITVAAFLAQQIRFTDIAALN
LSVLEKMDMREPQCVDDVLSVDANAREVARKEVMRLAS
|
|
|
|---|
| BDBM59101 |
|---|
| n/a |
|---|
| Name | BDBM59101 |
|---|
| Synonyms: | Bi-ligand, 4 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H15N3O8S3 |
|---|
| Mol. Mass. | 533.555 |
|---|
| SMILES | Oc1ccc(\C=C2/SC(=S)N(CC(=O)NCCSc3cc(nc(c3)C([O-])=O)C([O-])=O)C2=O)cc1O |
|---|
| Structure | 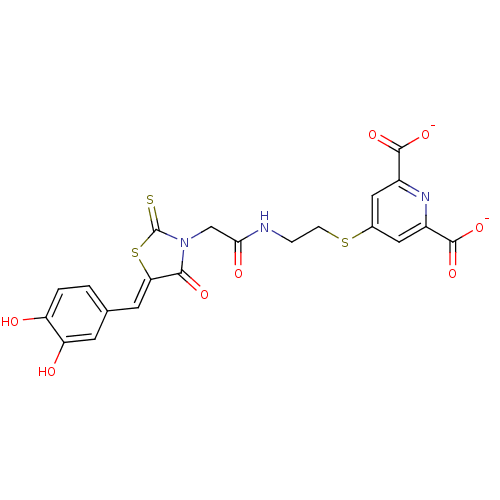
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Sem, DS; Bertolaet, B; Baker, B; Chang, E; Costache, AD; Coutts, S; Dong, Q; Hansen, M; Hong, V; Huang, X; Jack, RM; Kho, R; Lang, H; Ma, CT; Meininger, D; Pellecchia, M; Pierre, F; Villar, H; Yu, L Systems-based design of bi-ligand inhibitors of oxidoreductases: filling the chemical proteomic toolbox. Chem Biol11:185-94 (2004) [PubMed] Article
Sem, DS; Bertolaet, B; Baker, B; Chang, E; Costache, AD; Coutts, S; Dong, Q; Hansen, M; Hong, V; Huang, X; Jack, RM; Kho, R; Lang, H; Ma, CT; Meininger, D; Pellecchia, M; Pierre, F; Villar, H; Yu, L Systems-based design of bi-ligand inhibitors of oxidoreductases: filling the chemical proteomic toolbox. Chem Biol11:185-94 (2004) [PubMed] Article