| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase |
|---|
| Ligand | BDBM81626 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Inhibition Assay |
|---|
| IC50 | 2.5e+3±n/a nM |
|---|
| Citation |  Stamper, GF; Longenecker, KL; Fry, EH; Jakob, CG; Florjancic, AS; Gu, YG; Anderson, DD; Cooper, CS; Zhang, T; Clark, RF; Cia, Y; Black-Schaefer, CL; Owen McCall, J; Lerner, CG; Hajduk, PJ; Beutel, BA; Stoll, VS Structure-based optimization of MurF inhibitors. Chem Biol Drug Des67:58-65 (2006) [PubMed] Article Stamper, GF; Longenecker, KL; Fry, EH; Jakob, CG; Florjancic, AS; Gu, YG; Anderson, DD; Cooper, CS; Zhang, T; Clark, RF; Cia, Y; Black-Schaefer, CL; Owen McCall, J; Lerner, CG; Hajduk, PJ; Beutel, BA; Stoll, VS Structure-based optimization of MurF inhibitors. Chem Biol Drug Des67:58-65 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase |
|---|
| Name: | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase |
|---|
| Synonyms: | MurF (S. pneumoniae) | UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase (MurF) | UDP-N-acetylmuramoylalanine-D-glutamyl-lysine-D-alanyl-D-alanine ligase |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 50482.72 |
|---|
| Organism: | Streptococcus pneumoniae (Firmicutes) |
|---|
| Description: | S. pneumoniae MurF |
|---|
| Residue: | 457 |
|---|
| Sequence: | MKLTIHEIAQVVGAKNDISIFEDTQLEKAEFDSRLIGTGDLFVPLKGARDGHDFIETAFE
NGAAVTLSEKEVSNHPYILVDDVLTAFQSLASYYLEKTTVDVFAVTGSNGKTTTKDMLAH
LLSTRYKTYKTQGNYNNEIGLPYTVLHMPEGTEKLVLEMGQDHLGDIHLLSELARPKTAI
VTLVGEAHLAFFKDRSEIAKGKMQIADGMASGSLLLAPADPIVEDYLPIDKKVVRFGQGA
ELEITDLVERKDSLTFKANFLEQALDLPVTGKYNATNAMIASYVALQEGVSEEQIRLAFQ
HLELTRNRTEWKKAANGADILSDVYNANPTAMKLILETFSAIPANEGGKKIAVLADMKEL
GDQSVQLHNQMILSLSPDVLDIVIFYGEDIAQLAQLASQMFPIGHVYYFKKTEDQDQFED
LVKQVKESLGAHDQILLKGSNSMNLAKLVESLENEDK
|
|
|
|---|
| BDBM81626 |
|---|
| n/a |
|---|
| Name | BDBM81626 |
|---|
| Synonyms: | MurF inhibitor, 5 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H22ClN3O3S2 |
|---|
| Mol. Mass. | 464.001 |
|---|
| SMILES | Clc1ccc(cc1C(=O)Nc1sc2CCCCc2c1C#N)S(=O)(=O)N1CCCCC1 |
|---|
| Structure | 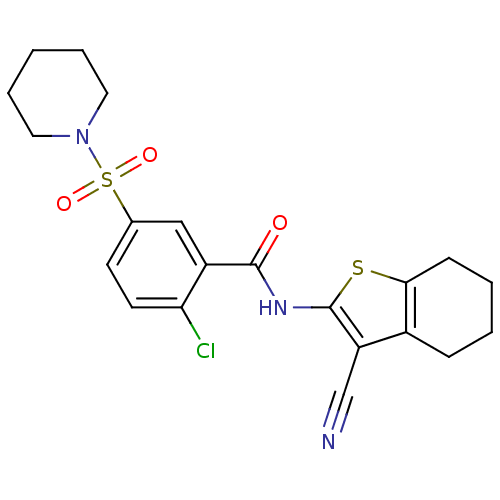
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Stamper, GF; Longenecker, KL; Fry, EH; Jakob, CG; Florjancic, AS; Gu, YG; Anderson, DD; Cooper, CS; Zhang, T; Clark, RF; Cia, Y; Black-Schaefer, CL; Owen McCall, J; Lerner, CG; Hajduk, PJ; Beutel, BA; Stoll, VS Structure-based optimization of MurF inhibitors. Chem Biol Drug Des67:58-65 (2006) [PubMed] Article
Stamper, GF; Longenecker, KL; Fry, EH; Jakob, CG; Florjancic, AS; Gu, YG; Anderson, DD; Cooper, CS; Zhang, T; Clark, RF; Cia, Y; Black-Schaefer, CL; Owen McCall, J; Lerner, CG; Hajduk, PJ; Beutel, BA; Stoll, VS Structure-based optimization of MurF inhibitors. Chem Biol Drug Des67:58-65 (2006) [PubMed] Article