| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Retinal dehydrogenase 2 |
|---|
| Ligand | BDBM50459586 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1766761 (CHEMBL4202008) |
|---|
| IC50 | 670±n/a nM |
|---|
| Citation |  Huddle, BC; Grimley, E; Buchman, CD; Chtcherbinine, M; Debnath, B; Mehta, P; Yang, K; Morgan, CA; Li, S; Felton, J; Sun, D; Mehta, G; Neamati, N; Buckanovich, RJ; Hurley, TD; Larsen, SD Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy. J Med Chem61:8754-8773 (2018) [PubMed] Article Huddle, BC; Grimley, E; Buchman, CD; Chtcherbinine, M; Debnath, B; Mehta, P; Yang, K; Morgan, CA; Li, S; Felton, J; Sun, D; Mehta, G; Neamati, N; Buckanovich, RJ; Hurley, TD; Larsen, SD Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy. J Med Chem61:8754-8773 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Retinal dehydrogenase 2 |
|---|
| Name: | Retinal dehydrogenase 2 |
|---|
| Synonyms: | AL1A2_HUMAN | ALDH1A2 | Aldehyde dehydrogenase family 1 member A2 | RALDH 2 | RALDH(II) | RALDH2 | Retinal dehydrogenase 2 | Retinaldehyde-specific dehydrogenase type 2 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 56720.77 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_109700 |
|---|
| Residue: | 518 |
|---|
| Sequence: | MTSSKIEMPGEVKADPAALMASLHLLPSPTPNLEIKYTKIFINNEWQNSESGRVFPVYNP
ATGEQVCEVQEADKADIDKAVQAARLAFSLGSVWRRMDASERGRLLDKLADLVERDRAVL
ATMESLNGGKPFLQAFYVDLQGVIKTFRYYAGWADKIHGMTIPVDGDYFTFTRHEPIGVC
GQIIPWNFPLLMFAWKIAPALCCGNTVVIKPAEQTPLSALYMGALIKEAGFPPGVINILP
GYGPTAGAAIASHIGIDKIAFTGSTEVGKLIQEAAGRSNLKRVTLELGGKSPNIIFADAD
LDYAVEQAHQGVFFNQGQCCTAGSRIFVEESIYEEFVRRSVERAKRRVVGSPFDPTTEQG
PQIDKKQYNKILELIQSGVAEGAKLECGGKGLGRKGFFIEPTVFSNVTDDMRIAKEEIFG
PVQEILRFKTMDEVIERANNSDFGLVAAVFTNDINKALTVSSAMQAGTVWINCYNALNAQ
SPFGGFKMSGNGREMGEFGLREYSEVKTVTVKIPQKNS
|
|
|
|---|
| BDBM50459586 |
|---|
| n/a |
|---|
| Name | BDBM50459586 |
|---|
| Synonyms: | CHEMBL4217294 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H13FN4OS |
|---|
| Mol. Mass. | 352.385 |
|---|
| SMILES | Fc1cccc(CSc2nc3[nH]ncc3c(=O)n2-c2ccccc2)c1 |
|---|
| Structure | 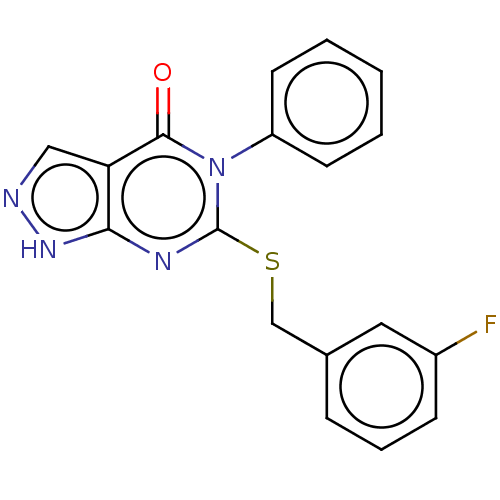
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Huddle, BC; Grimley, E; Buchman, CD; Chtcherbinine, M; Debnath, B; Mehta, P; Yang, K; Morgan, CA; Li, S; Felton, J; Sun, D; Mehta, G; Neamati, N; Buckanovich, RJ; Hurley, TD; Larsen, SD Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy. J Med Chem61:8754-8773 (2018) [PubMed] Article
Huddle, BC; Grimley, E; Buchman, CD; Chtcherbinine, M; Debnath, B; Mehta, P; Yang, K; Morgan, CA; Li, S; Felton, J; Sun, D; Mehta, G; Neamati, N; Buckanovich, RJ; Hurley, TD; Larsen, SD Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy. J Med Chem61:8754-8773 (2018) [PubMed] Article