| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacetylase sirtuin-3, mitochondrial |
|---|
| Ligand | BDBM50459842 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1767310 (CHEMBL4219422) |
|---|
| IC50 | >100000±n/a nM |
|---|
| Citation |  Schiedel, M; Herp, D; Hammelmann, S; Swyter, S; Lehotzky, A; Robaa, D; Oláh, J; Ovádi, J; Sippl, W; Jung, M Chemically Induced Degradation of Sirtuin 2 (Sirt2) by a Proteolysis Targeting Chimera (PROTAC) Based on Sirtuin Rearranging Ligands (SirReals). J Med Chem61:482-491 (2018) [PubMed] Article Schiedel, M; Herp, D; Hammelmann, S; Swyter, S; Lehotzky, A; Robaa, D; Oláh, J; Ovádi, J; Sippl, W; Jung, M Chemically Induced Degradation of Sirtuin 2 (Sirt2) by a Proteolysis Targeting Chimera (PROTAC) Based on Sirtuin Rearranging Ligands (SirReals). J Med Chem61:482-491 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacetylase sirtuin-3, mitochondrial |
|---|
| Name: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial |
|---|
| Synonyms: | NAD-dependent deacetylase sirtuin 3 | NAD-dependent protein deacetylase sirtuin-3 (SIRT3) | SIR2L3 | SIR3_HUMAN | SIRT3 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 43582.69 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9NTG7 |
|---|
| Residue: | 399 |
|---|
| Sequence: | MAFWGWRAAAALRLWGRVVERVEAGGGVGPFQACGCRLVLGGRDDVSAGLRGSHGARGEP
LDPARPLQRPPRPEVPRAFRRQPRAAAPSFFFSSIKGGRRSISFSVGASSVVGSGGSSDK
GKLSLQDVAELIRARACQRVVVMVGAGISTPSGIPDFRSPGSGLYSNLQQYDLPYPEAIF
ELPFFFHNPKPFFTLAKELYPGNYKPNVTHYFLRLLHDKGLLLRLYTQNIDGLERVSGIP
ASKLVEAHGTFASATCTVCQRPFPGEDIRADVMADRVPRCPVCTGVVKPDIVFFGEPLPQ
RFLLHVVDFPMADLLLILGTSLEVEPFASLTEAVRSSVPRLLINRDLVGPLAWHPRSRDV
AQLGDVVHGVESLVELLGWTEEMRDLVQRETGKLDGPDK
|
|
|
|---|
| BDBM50459842 |
|---|
| n/a |
|---|
| Name | BDBM50459842 |
|---|
| Synonyms: | CHEMBL4228286 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H27N7O2S2 |
|---|
| Mol. Mass. | 557.69 |
|---|
| SMILES | Cc1cc(C)nc(SCC(=O)Nc2ncc(Cc3cccc(OCc4cn(Cc5ccccc5)nn4)c3)s2)n1 |
|---|
| Structure | 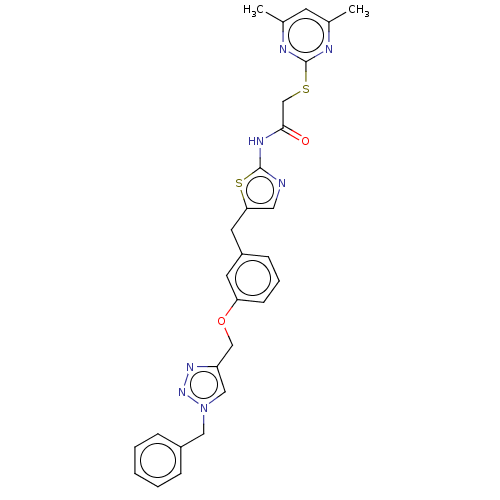
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Schiedel, M; Herp, D; Hammelmann, S; Swyter, S; Lehotzky, A; Robaa, D; Oláh, J; Ovádi, J; Sippl, W; Jung, M Chemically Induced Degradation of Sirtuin 2 (Sirt2) by a Proteolysis Targeting Chimera (PROTAC) Based on Sirtuin Rearranging Ligands (SirReals). J Med Chem61:482-491 (2018) [PubMed] Article
Schiedel, M; Herp, D; Hammelmann, S; Swyter, S; Lehotzky, A; Robaa, D; Oláh, J; Ovádi, J; Sippl, W; Jung, M Chemically Induced Degradation of Sirtuin 2 (Sirt2) by a Proteolysis Targeting Chimera (PROTAC) Based on Sirtuin Rearranging Ligands (SirReals). J Med Chem61:482-491 (2018) [PubMed] Article