| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Genome polyprotein |
|---|
| Ligand | BDBM50106501 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1772460 (CHEMBL4224572) |
|---|
| IC50 | 630±n/a nM |
|---|
| Citation |  Jung, E; Lee, JY; Kim, HJ; Ryu, CK; Lee, KI; Kim, M; Lee, CK; Go, YY Identification of quinone analogues as potential inhibitors of picornavirus 3C protease in vitro. Bioorg Med Chem Lett28:2533-2538 (2018) [PubMed] Article Jung, E; Lee, JY; Kim, HJ; Ryu, CK; Lee, KI; Kim, M; Lee, CK; Go, YY Identification of quinone analogues as potential inhibitors of picornavirus 3C protease in vitro. Bioorg Med Chem Lett28:2533-2538 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Genome polyprotein |
|---|
| Name: | Genome polyprotein |
|---|
| Synonyms: | 3C-like proteinase (CVB3 3Cpro) | Genome polyprotein | P1A | P1B | P1C | P1D | P2A | P2B | P2C | P3A | P3B | P3C | P3D-POL | POLG_CXB3N | Picornain 3C | Polyprotein | Protease 2A | Protease 3C | Protein 2A | Protein 2B | Protein 2C | Protein 3A | Protein 3B | Protein VP0 | Protein VP1 | Protein VP2 | Protein VP3 | Protein VP4 | RNA-directed RNA polymerase 3D-POL | VP4-VP2 | VPg | Virion protein 1 | Virion protein 2 | Virion protein 3 | Virion protein 4 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 243464.96 |
|---|
| Organism: | Coxsackievirus B3 (strain Nancy) |
|---|
| Description: | P03313 |
|---|
| Residue: | 2185 |
|---|
| Sequence: | MGAQVSTQKTGAHETRLNASGNSIIHYTNINYYKDAASNSANRQDFTQDPGKFTEPVKDI
MIKSLPALNSPTVEECGYSDRARSITLGNSTITTQECANVVVGYGVWPDYLKDSEATAED
QPTQPDVATCRFYTLDSVQWQKTSPGWWWKLPDALSNLGLFGQNMQYHYLGRTGYTVHVQ
CNASKFHQGCLLVVCVPEAEMGCATLDNTPSSAELLGGDTAKEFADKPVASGSNKLVQRV
VYNAGMGVGVGNLTIFPHQWINLRTNNSATIVMPYTNSVPMDNMFRHNNVTLMVIPFVPL
DYCPGSTTYVPITVTIAPMCAEYNGLRLAGHQGLPTMNTPGSCQFLTSDDFQSPSAMPQY
DVTPEMRIPGEVKNLMEIAEVDSVVPVQNVGEKVNSMEAYQIPVRSNEGSGTQVFGFPLQ
PGYSSVFSRTLLGEILNYYTHWSGSIKLTFMFCGSAMATGKFLLAYSPPGAGAPTKRVDA
MLGTHVIWDVGLQSSCVLCIPWISQTHYRFVASDEYTAGGFITCWYQTNIVVPADAQSSC
YIMCFVSACNDFSVRLLKDTPFISQQNFFQGPVEDAITAAIGRVADTVGTGPTNSEAIPA
LTAAETGHTSQVVPGDTMQTRHVKNYHSRSESTIENFLCRSACVYFTEYKNSGAKRYAEW
VLTPRQAAQLRRKLEFFTYVRFDLELTFVITSTQQPSTTQNQDAQILTHQIMYVPPGGPV
PDKVDSYVWQTSTNPSVFWTEGNAPPRMSIPFLSIGNAYSNFYDGWSEFSRNGVYGINTL
NNMGTLYARHVNAGSTGPIKSTIRIYFKPKHVKAWIPRPPRLCQYEKAKNVNFQPSGVTT
TRQSITTMTNTGAFGQQSGAVYVGNYRVVNRHLATSADWQNCVWESYNRDLLVSTTTAHG
CDIIARCQCTTGVYFCASKNKHYPISFEGPGLVEVQESEYYPRRYQSHVLLAAGFSEPGD
CGGILRCEHGVIGIVTMGGEGVVGFADIRDLLWLEDDAMEQGVKDYVEQLGNAFGSGFTN
QICEQVNLLKESLVGQDSILEKSLKALVKIISALVIVVRNHDDLITVTATLALIGCTSSP
WRWLKQKVSQYYGIPMAERQNNSWLKKFTEMTNACKGMEWIAVKIQKFIEWLKVKILPEV
REKHEFLNRLKQLPLLESQIATIEQSAPSQSDQEQLFSNVQYFAHYCRKYAPLYAAEAKR
VFSLEKKMSNYIQFKSKCRIEPVCLLLHGSPGAGKSVATNLIGRSLAEKLNSSVYSLPPD
PDHFDGYKQQAVVIMDDLCQNPDGKDVSLFCQMVSSVDFVPPMAALEEKGILFTSPFVLA
STNAGSINAPTVSDSRALARRFHFDMNIEVISMYSQNGKINMPMSVKTCDDECCPVNFKK
CCPLVCGKAIQFIDRRTQVRYSLDMLVTEMFREYNHRHSVGTTLEALFQGPPVYREIKIS
VAPETPPPPAIADLLKSVDSEAVREYCKEKGWLVPEINSTLQIEKHVSRAFICLQALTTF
VSVAGIIYIIYKLFAGFQGAYTGVPNQKPRVPTLRQAKVQGPAFEFAVAMMKRNSSTVKT
EYGEFTMLGIYDRWAVLPRHAKPGPTILMNDQEVGVLDAKELVDKDGTNLELTLLKLNRN
EKFRDIRGFLAKEEVEVNEAVLAINTSKFPNMYIPVGQVTEYGFLNLGGTPTKRMLMYNF
PTRAGQCGGVLMSTGKVLGIHVGGNGHQGFSAALLKHYFNDEQGEIEFIESSKDAGFPVI
NTPSKTKLEPSVFHQVFEGNKEPAVLRSGDPRLKANFEEAIFSKYIGNVNTHVDEYMLEA
VDHYAGQLATLDISTEPMKLEDAVYGTEGLEALDLTTSAGYPYVALGIKKRDILSKKTKD
LTKLKECMDKYGLNLPMVTYVKDELRSIEKVAKGKSRLIEASSLNDSVAMRQTFGNLYKT
FHLNPGVVTGSAVGCDPDLFWSKIPVMLDGHLIAFDYSGYDASLSPVWFACLKMLLEKLG
YTHKETNYIDYLCNSHHLYRDKHYFVRGGMPSGCSGTSIFNSMINNIIIRTLMLKVYKGI
DLDQFRMIAYGDDVIASYPWPIDASLLAEAGKGYGLIMTPADKGECFNEVTWTNATFLKR
YFRADEQYPFLVHPVMPMKDIHESIRWTKDPKNTQDHVRSLCLLAWHNGEHEYEEFIRKI
RSVPVGRCLTLPAFSTLRRKWLDSF
|
|
|
|---|
| BDBM50106501 |
|---|
| n/a |
|---|
| Name | BDBM50106501 |
|---|
| Synonyms: | 6,7-Dichloro-5,8-quinolinequinone | 6,7-Dichloro-quinoline-5,8-dione | CHEMBL132160 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C9H3Cl2NO2 |
|---|
| Mol. Mass. | 228.032 |
|---|
| SMILES | ClC1=C(Cl)C(=O)c2ncccc2C1=O |c:1| |
|---|
| Structure | 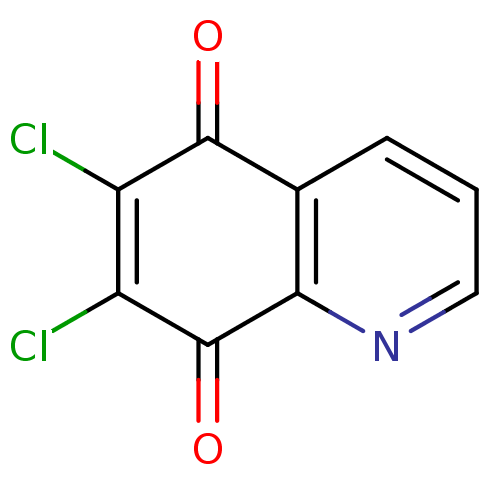
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jung, E; Lee, JY; Kim, HJ; Ryu, CK; Lee, KI; Kim, M; Lee, CK; Go, YY Identification of quinone analogues as potential inhibitors of picornavirus 3C protease in vitro. Bioorg Med Chem Lett28:2533-2538 (2018) [PubMed] Article
Jung, E; Lee, JY; Kim, HJ; Ryu, CK; Lee, KI; Kim, M; Lee, CK; Go, YY Identification of quinone analogues as potential inhibitors of picornavirus 3C protease in vitro. Bioorg Med Chem Lett28:2533-2538 (2018) [PubMed] Article