| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Ligand | BDBM50484893 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_810002 (CHEMBL2015111) |
|---|
| IC50 | 26±n/a nM |
|---|
| Citation |  Warmus, JS; Quinn, CL; Taylor, C; Murphy, ST; Johnson, TA; Limberakis, C; Ortwine, D; Bronstein, J; Pagano, P; Knafels, JD; Lightle, S; Mochalkin, I; Brideau, R; Podoll, T Structure based design of an in vivo active hydroxamic acid inhibitor of P. aeruginosa LpxC. Bioorg Med Chem Lett22:2536-43 (2012) [PubMed] Article Warmus, JS; Quinn, CL; Taylor, C; Murphy, ST; Johnson, TA; Limberakis, C; Ortwine, D; Bronstein, J; Pagano, P; Knafels, JD; Lightle, S; Mochalkin, I; Brideau, R; Podoll, T Structure based design of an in vivo active hydroxamic acid inhibitor of P. aeruginosa LpxC. Bioorg Med Chem Lett22:2536-43 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Name: | UDP-3-O-acyl-N-acetylglucosamine deacetylase |
|---|
| Synonyms: | LPXC_PSEAE | Protein envA | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LpxC | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LpxC) | UDP-3-O-acyl-GlcNAc deacetylase | UDP-3-O-acyl-GlcNAc deacetylase (LpxC) | envA | lpxC |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 33428.15 |
|---|
| Organism: | Pseudomonas aeruginosa |
|---|
| Description: | P47205 |
|---|
| Residue: | 303 |
|---|
| Sequence: | MIKQRTLKNIIRATGVGLHSGEKVYLTLKPAPVDTGIVFCRTDLDPVVEIPARAENVGET
TMSTTLVKGDVKVDTVEHLLSAMAGLGIDNAYVELSASEVPIMDGSAGPFVFLIQSAGLQ
EQEAAKKFIRIKREVSVEEGDKRAVFVPFDGFKVSFEIDFDHPVFRGRTQQASVDFSSTS
FVKEVSRARTFGFMRDIEYLRSQNLALGGSVENAIVVDENRVLNEDGLRYEDEFVKHKIL
DAIGDLYLLGNSLIGEFRGFKSGHALNNQLLRTLIADKDAWEVVTFEDARTAPISYMRPA
AAV
|
|
|
|---|
| BDBM50484893 |
|---|
| n/a |
|---|
| Name | BDBM50484893 |
|---|
| Synonyms: | CHEMBL2012197 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H22N4O4 |
|---|
| Mol. Mass. | 382.4131 |
|---|
| SMILES | Cn1nncc1[C@H](O)[C@](C)(OCc1ccc(cc1)-c1ccccc1)C(=O)NO |r| |
|---|
| Structure | 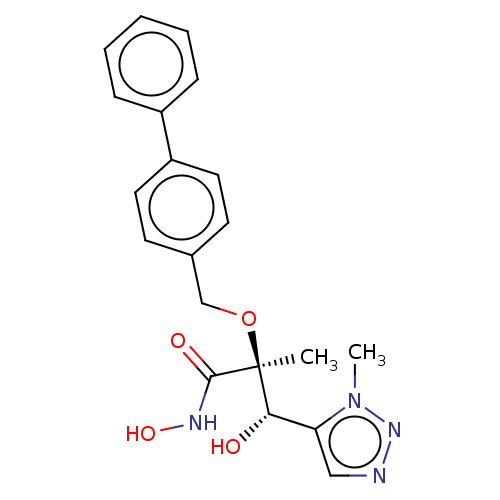
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Warmus, JS; Quinn, CL; Taylor, C; Murphy, ST; Johnson, TA; Limberakis, C; Ortwine, D; Bronstein, J; Pagano, P; Knafels, JD; Lightle, S; Mochalkin, I; Brideau, R; Podoll, T Structure based design of an in vivo active hydroxamic acid inhibitor of P. aeruginosa LpxC. Bioorg Med Chem Lett22:2536-43 (2012) [PubMed] Article
Warmus, JS; Quinn, CL; Taylor, C; Murphy, ST; Johnson, TA; Limberakis, C; Ortwine, D; Bronstein, J; Pagano, P; Knafels, JD; Lightle, S; Mochalkin, I; Brideau, R; Podoll, T Structure based design of an in vivo active hydroxamic acid inhibitor of P. aeruginosa LpxC. Bioorg Med Chem Lett22:2536-43 (2012) [PubMed] Article