| Citation |  Gong, L; Han, X; Silva, T; Tan, YC; Goyal, B; Tivitmahaisoon, P; Trejo, A; Palmer, W; Hogg, H; Jahagir, A; Alam, M; Wagner, P; Stein, K; Filonova, L; Loe, B; Makra, F; Rotstein, D; Rapatova, L; Dunn, J; Zuo, F; Dal Porto, J; Wong, B; Jin, S; Chang, A; Tran, P; Hsieh, G; Niu, L; Shao, A; Reuter, D; Hermann, J; Kuglstatter, A; Goldstein, D Development of indole/indazole-aminopyrimidines as inhibitors of c-Jun N-terminal kinase (JNK): optimization for JNK potency and physicochemical properties. Bioorg Med Chem Lett23:3565-9 (2013) [PubMed] Article Gong, L; Han, X; Silva, T; Tan, YC; Goyal, B; Tivitmahaisoon, P; Trejo, A; Palmer, W; Hogg, H; Jahagir, A; Alam, M; Wagner, P; Stein, K; Filonova, L; Loe, B; Makra, F; Rotstein, D; Rapatova, L; Dunn, J; Zuo, F; Dal Porto, J; Wong, B; Jin, S; Chang, A; Tran, P; Hsieh, G; Niu, L; Shao, A; Reuter, D; Hermann, J; Kuglstatter, A; Goldstein, D Development of indole/indazole-aminopyrimidines as inhibitors of c-Jun N-terminal kinase (JNK): optimization for JNK potency and physicochemical properties. Bioorg Med Chem Lett23:3565-9 (2013) [PubMed] Article |
|---|
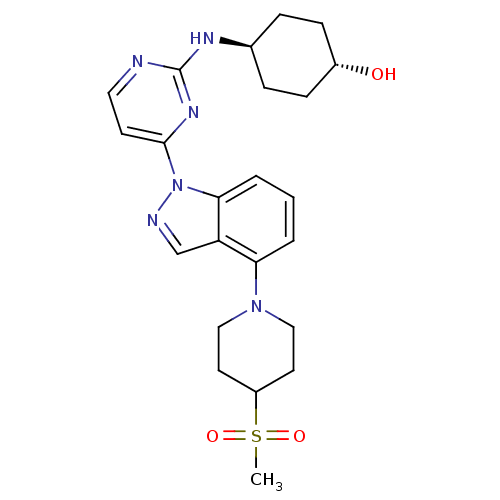
| SMILES | CS(=O)(=O)C1CCN(CC1)c1cccc2n(ncc12)-c1ccnc(N[C@H]2CC[C@H](O)CC2)n1 |r,wU:25.27,wD:28.31,(10.53,3.6,;9.19,2.83,;8.42,4.17,;9.96,1.5,;7.86,2.06,;7.86,.52,;6.52,-.25,;5.19,.52,;5.19,2.06,;6.52,2.83,;3.86,-.25,;3.86,-1.79,;2.52,-2.56,;1.19,-1.79,;1.19,-.25,;.04,.78,;.67,2.19,;2.2,2.03,;2.52,.52,;-1.5,.78,;-2.27,2.12,;-3.81,2.12,;-4.58,.78,;-3.81,-.55,;-4.58,-1.88,;-6.06,-2.28,;-6.46,-3.77,;-7.95,-4.17,;-9.04,-3.08,;-10.53,-3.48,;-8.64,-1.59,;-7.15,-1.19,;-2.27,-.55,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Gong, L; Han, X; Silva, T; Tan, YC; Goyal, B; Tivitmahaisoon, P; Trejo, A; Palmer, W; Hogg, H; Jahagir, A; Alam, M; Wagner, P; Stein, K; Filonova, L; Loe, B; Makra, F; Rotstein, D; Rapatova, L; Dunn, J; Zuo, F; Dal Porto, J; Wong, B; Jin, S; Chang, A; Tran, P; Hsieh, G; Niu, L; Shao, A; Reuter, D; Hermann, J; Kuglstatter, A; Goldstein, D Development of indole/indazole-aminopyrimidines as inhibitors of c-Jun N-terminal kinase (JNK): optimization for JNK potency and physicochemical properties. Bioorg Med Chem Lett23:3565-9 (2013) [PubMed] Article
Gong, L; Han, X; Silva, T; Tan, YC; Goyal, B; Tivitmahaisoon, P; Trejo, A; Palmer, W; Hogg, H; Jahagir, A; Alam, M; Wagner, P; Stein, K; Filonova, L; Loe, B; Makra, F; Rotstein, D; Rapatova, L; Dunn, J; Zuo, F; Dal Porto, J; Wong, B; Jin, S; Chang, A; Tran, P; Hsieh, G; Niu, L; Shao, A; Reuter, D; Hermann, J; Kuglstatter, A; Goldstein, D Development of indole/indazole-aminopyrimidines as inhibitors of c-Jun N-terminal kinase (JNK): optimization for JNK potency and physicochemical properties. Bioorg Med Chem Lett23:3565-9 (2013) [PubMed] Article