

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Sterol O-acyltransferase 1 | ||
| Ligand | BDBM50051890 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_28658 (CHEMBL643993) | ||
| IC50 | 60±n/a nM | ||
| Citation |  Astles, PC; Ashton, MJ; Bridge, AW; Harris, NV; Hart, TW; Parrott, DP; Porter, B; Riddell, D; Smith, C; Williams, RJ Acyl-CoA:Cholesterol O-acyltransferase (ACAT) inhibitors. 2. 2-(1,3-Dioxan-2-yl)-4,5-diphenyl-1H-imidazoles as potent inhibitors of ACAT. J Med Chem39:1423-32 (1996) [PubMed] Article Astles, PC; Ashton, MJ; Bridge, AW; Harris, NV; Hart, TW; Parrott, DP; Porter, B; Riddell, D; Smith, C; Williams, RJ Acyl-CoA:Cholesterol O-acyltransferase (ACAT) inhibitors. 2. 2-(1,3-Dioxan-2-yl)-4,5-diphenyl-1H-imidazoles as potent inhibitors of ACAT. J Med Chem39:1423-32 (1996) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Sterol O-acyltransferase 1 | |||
| Name: | Sterol O-acyltransferase 1 | ||
| Synonyms: | ACACT | ACACT1 | ACAT | ACAT1 | Acetyl-CoA acetyltransferase, mitochondrial | Acyl coenzyme A:cholesterol acyltransferase 1 | Acyl-CoA: cholesterol acyltransferase (ACAT) | Acyl-coenzyme A:cholesterol acyltransferase 1 (ACAT1) | Cholesterol acyltransferase 1 | SOAT | SOAT1 | SOAT1_HUMAN | STAT | ||
| Type: | Multi-pass membrane protein may form homo- or heterodimers. | ||
| Mol. Mass.: | 64751.94 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P35610 | ||
| Residue: | 550 | ||
| Sequence: |
| ||
| BDBM50051890 | |||
| n/a | |||
| Name | BDBM50051890 | ||
| Synonyms: | 2-{4-[2-(4,5-Diphenyl-1H-imidazol-2-yl)-5-methyl-[1,3]dioxan-5-yl]-butyl}-1,2,3,4-tetrahydro-isoquinoline | CHEMBL33054 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C33H37N3O2 | ||
| Mol. Mass. | 507.6658 | ||
| SMILES | CC1(CCCCN2CCc3ccccc3C2)COC(OC1)c1nc(c([nH]1)-c1ccccc1)-c1ccccc1 |(11.65,-14.64,;10.9,-13.31,;12.1,-12.35,;13.52,-12.87,;14.71,-11.9,;16.16,-12.44,;17.36,-11.46,;17.35,-9.92,;18.69,-9.15,;20.02,-9.92,;21.33,-9.17,;22.66,-9.95,;22.66,-11.46,;21.32,-12.23,;20,-11.46,;18.69,-12.24,;9.62,-13.73,;7.98,-13.12,;7.22,-14.47,;8.4,-14.05,;10.13,-14.66,;5.88,-13.69,;5.47,-12.19,;3.93,-12.12,;3.37,-13.57,;4.57,-14.54,;1.88,-13.97,;.8,-12.88,;-.69,-13.29,;-1.09,-14.79,;.01,-15.86,;1.5,-15.46,;3.07,-10.83,;3.77,-9.45,;2.93,-8.17,;1.39,-8.24,;.7,-9.63,;1.54,-10.92,)| | ||
| Structure | 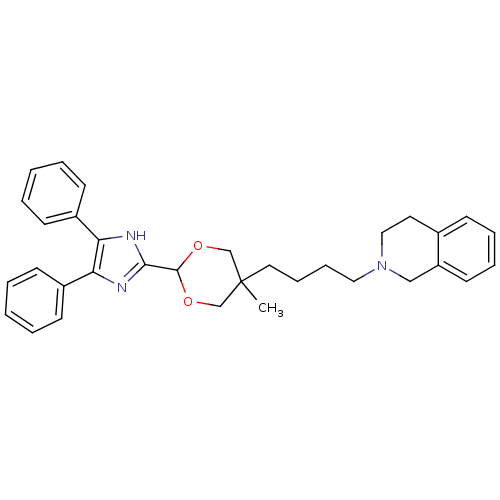
| ||