

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | D(3) dopamine receptor | ||
| Ligand | BDBM50052181 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_62756 (CHEMBL674596) | ||
| Ki | 1.7±n/a nM | ||
| Citation |  Ohmori, J; Maeno, K; Hidaka, K; Nakato, K; Matsumoto, M; Tada, S; Hattori, H; Sakamoto, S; Tsukamoto, S; Usuda, S; Mase, T Dopamine D3 and D4 receptor antagonists: synthesis and structure--activity relationships of (S)-(+)-N-(1-Benzyl-3-pyrrolidinyl)-5-chloro-4- [(cyclopropylcarbonyl) amino]-2-methoxybenzamide (YM-43611) and related compounds. J Med Chem39:2764-72 (1996) [PubMed] Article Ohmori, J; Maeno, K; Hidaka, K; Nakato, K; Matsumoto, M; Tada, S; Hattori, H; Sakamoto, S; Tsukamoto, S; Usuda, S; Mase, T Dopamine D3 and D4 receptor antagonists: synthesis and structure--activity relationships of (S)-(+)-N-(1-Benzyl-3-pyrrolidinyl)-5-chloro-4- [(cyclopropylcarbonyl) amino]-2-methoxybenzamide (YM-43611) and related compounds. J Med Chem39:2764-72 (1996) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| D(3) dopamine receptor | |||
| Name: | D(3) dopamine receptor | ||
| Synonyms: | DOPAMINE D3 | DRD3_RAT | Dopamine D3 receptor | Drd3 | ||
| Type: | Protein | ||
| Mol. Mass.: | 49540.58 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | P19020 | ||
| Residue: | 446 | ||
| Sequence: |
| ||
| BDBM50052181 | |||
| n/a | |||
| Name | BDBM50052181 | ||
| Synonyms: | CHEMBL92924 | N-((S)-1-Adamantan-2-yl-pyrrolidin-3-yl)-5-chloro-4-(cyclopropanecarbonyl-amino)-2-methoxy-benzamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H34ClN3O3 | ||
| Mol. Mass. | 472.019 | ||
| SMILES | COc1cc(NC(=O)C2CC2)c(Cl)cc1C(=O)N[C@H]1CCN(C1)C1C2CC3CC(C2)CC1C3 |wU:18.19,TLB:29:24:32:28.30.27,29:28:23.24.25:32,THB:21:23:32:28.30.27,27:28:23:26.25.32,27:26:23:28.30.29,(5.91,-4.27,;5.93,-2.73,;4.6,-1.96,;3.25,-2.73,;1.92,-1.96,;.59,-2.73,;-.74,-1.96,;-.74,-.4,;-2.08,-2.73,;-3.62,-2.73,;-2.85,-4.06,;1.92,-.4,;.59,.35,;3.25,.37,;4.6,-.4,;5.93,.37,;5.93,1.91,;7.23,-.4,;8.34,.69,;8.34,2.23,;11,2.24,;11.07,.74,;9.68,-.08,;12.42,,;13.77,.48,;12.72,-.75,;12.72,-2.34,;14.14,-2.9,;15.15,-1.62,;15.15,-.1,;13.75,-1.97,;12.42,-1.48,;11.23,-2.76,)| | ||
| Structure | 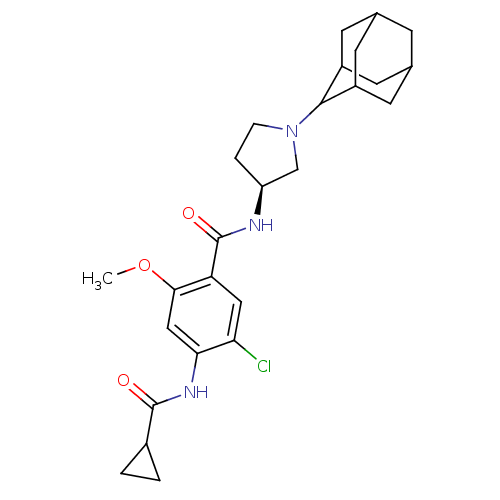
| ||