

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Peroxisome proliferator-activated receptor gamma | ||
| Ligand | BDBM50519893 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1878653 (CHEMBL4380047) | ||
| IC50 | >50000±n/a nM | ||
| Citation |  Meijer, FA; Doveston, RG; de Vries, RMJM; Vos, GM; Vos, AAA; Leysen, S; Scheepstra, M; Ottmann, C; Milroy, LG; Brunsveld, L Ligand-Based Design of Allosteric Retinoic Acid Receptor-Related Orphan Receptor ?t (ROR?t) Inverse Agonists. J Med Chem63:241-259 (2020) [PubMed] Article Meijer, FA; Doveston, RG; de Vries, RMJM; Vos, GM; Vos, AAA; Leysen, S; Scheepstra, M; Ottmann, C; Milroy, LG; Brunsveld, L Ligand-Based Design of Allosteric Retinoic Acid Receptor-Related Orphan Receptor ?t (ROR?t) Inverse Agonists. J Med Chem63:241-259 (2020) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Peroxisome proliferator-activated receptor gamma | |||
| Name: | Peroxisome proliferator-activated receptor gamma | ||
| Synonyms: | NR1C3 | Nuclear receptor subfamily 1 group C member 3 | PPAR-gamma | PPARG | PPARG_HUMAN | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor gamma (PPAR gamma) | Peroxisome proliferator-activated receptor gamma (PPARG) | Peroxisome proliferator-activated receptor gamma (PPARγ) | Peroxisome proliferator-activated receptor gamma/Nuclear receptor corepressor 2 | peroxisome proliferator-activated receptor gamma isoform 2 | ||
| Type: | Nuclear Receptor | ||
| Mol. Mass.: | 57613.46 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P37231 | ||
| Residue: | 505 | ||
| Sequence: |
| ||
| BDBM50519893 | |||
| n/a | |||
| Name | BDBM50519893 | ||
| Synonyms: | CHEMBL4437814 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H14ClF3N2O5S | ||
| Mol. Mass. | 522.881 | ||
| SMILES | OC(=O)c1ccc(cc1)S(=O)(=O)Nc1c(noc1-c1ccccc1)-c1c(Cl)cccc1C(F)(F)F |(19.75,-25.52,;18.61,-26.56,;18.95,-28.06,;17.15,-26.1,;16.01,-27.14,;14.54,-26.67,;14.22,-25.17,;15.35,-24.13,;16.81,-24.59,;12.75,-24.69,;11.73,-23.52,;13.26,-23.23,;11.62,-25.74,;10.15,-25.27,;8.91,-26.18,;7.65,-25.29,;8.11,-23.81,;9.66,-23.8,;10.56,-22.56,;12.09,-22.71,;12.98,-21.47,;12.35,-20.06,;10.81,-19.91,;9.92,-21.16,;8.93,-27.77,;10.3,-28.53,;11.66,-27.72,;10.33,-30.1,;8.97,-30.92,;7.59,-30.14,;7.57,-28.57,;6.23,-27.81,;6.22,-26.27,;4.91,-28.59,;4.89,-27.04,)| | ||
| Structure | 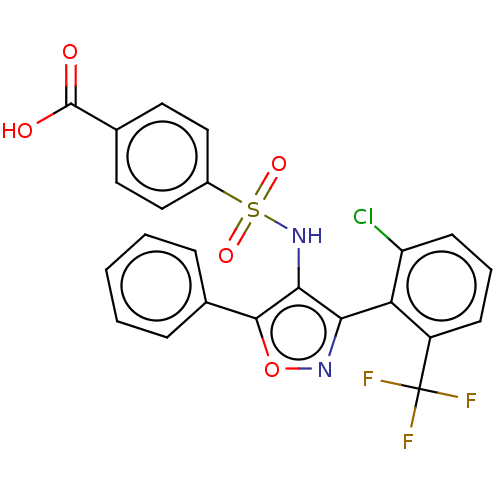
| ||