| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Low affinity immunoglobulin epsilon Fc receptor |
|---|
| Ligand | BDBM50082229 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_88888 |
|---|
| IC50 | 230±n/a nM |
|---|
| Citation |  Bailey, S; Bolognese, B; Faller, A; Louis-Flamberg, P; MacPherson, DT; Mayer, RJ; Marshall, LA; Milner, PH; Mistry, J; Smith, DG; Ward, JG Selective inhibition of low affinity IgE receptor (CD23) processing: P1' bicyclomethyl substituents. Bioorg Med Chem Lett9:3165-70 (1999) [PubMed] Bailey, S; Bolognese, B; Faller, A; Louis-Flamberg, P; MacPherson, DT; Mayer, RJ; Marshall, LA; Milner, PH; Mistry, J; Smith, DG; Ward, JG Selective inhibition of low affinity IgE receptor (CD23) processing: P1' bicyclomethyl substituents. Bioorg Med Chem Lett9:3165-70 (1999) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Low affinity immunoglobulin epsilon Fc receptor |
|---|
| Name: | Low affinity immunoglobulin epsilon Fc receptor |
|---|
| Synonyms: | BLAST-2 | CD23A | CD_antigen=CD23 | CLEC4J | FCE2 | FCER2 | FCER2_HUMAN | Fc-epsilon-RII | IGEBF | Immunoglobulin E-binding factor | Immunoglobulin epsilon Fc receptor | Low affinity immunoglobulin epsilon Fc receptor | Low affinity immunoglobulin epsilon Fc receptor membrane-bound form | Low affinity immunoglobulin epsilon Fc receptor soluble form | Lymphocyte IgE receptor |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 36461.84 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_88888 |
|---|
| Residue: | 321 |
|---|
| Sequence: | MEEGQYSEIEELPRRRCCRRGTQIVLLGLVTAALWAGLLTLLLLWHWDTTQSLKQLEERA
ARNVSQVSKNLESHHGDQMAQKSQSTQISQELEELRAEQQRLKSQDLELSWNLNGLQADL
SSFKSQELNERNEASDLLERLREEVTKLRMELQVSSGFVCNTCPEKWINFQRKCYYFGKG
TKQWVHARYACDDMEGQLVSIHSPEEQDFLTKHASHTGSWIGLRNLDLKGEFIWVDGSHV
DYSNWAPGEPTSRSQGEDCVMMRGSGRWNDAFCDRKLGAWVCDRLATCTPPASEGSAESM
GPDSRPDPDGRLPTPSAPLHS
|
|
|
|---|
| BDBM50082229 |
|---|
| n/a |
|---|
| Name | BDBM50082229 |
|---|
| Synonyms: | (2R,3S)-N*4*-((S)-2,2-Dimethyl-1-methylcarbamoyl-propyl)-2,N*1*-dihydroxy-3-naphthalen-2-ylmethyl-succinamide | CHEMBL108737 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H29N3O5 |
|---|
| Mol. Mass. | 415.4828 |
|---|
| SMILES | CNC(=O)[C@@H](NC(=O)[C@H](Cc1ccc2ccccc2c1)[C@H](O)C(=O)NO)C(C)(C)C |
|---|
| Structure | 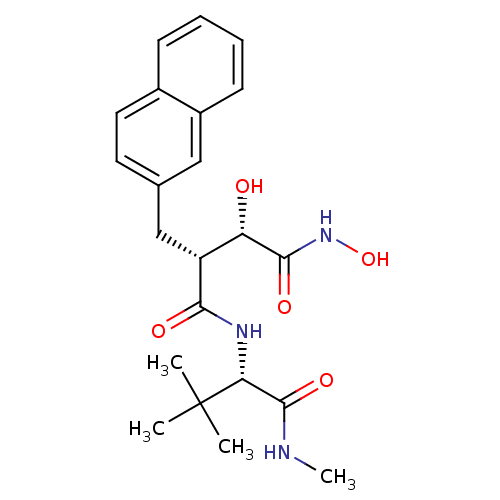
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bailey, S; Bolognese, B; Faller, A; Louis-Flamberg, P; MacPherson, DT; Mayer, RJ; Marshall, LA; Milner, PH; Mistry, J; Smith, DG; Ward, JG Selective inhibition of low affinity IgE receptor (CD23) processing: P1' bicyclomethyl substituents. Bioorg Med Chem Lett9:3165-70 (1999) [PubMed]
Bailey, S; Bolognese, B; Faller, A; Louis-Flamberg, P; MacPherson, DT; Mayer, RJ; Marshall, LA; Milner, PH; Mistry, J; Smith, DG; Ward, JG Selective inhibition of low affinity IgE receptor (CD23) processing: P1' bicyclomethyl substituents. Bioorg Med Chem Lett9:3165-70 (1999) [PubMed]