

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 3A4 | ||
| Ligand | BDBM50512456 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1924446 (CHEMBL4427402) | ||
| IC50 | 14000±n/a nM | ||
| Citation |  Liu, Y; Laufer, R; Patel, NK; Ng, G; Sampson, PB; Li, SW; Lang, Y; Feher, M; Brokx, R; Beletskaya, I; Hodgson, R; Plotnikova, O; Awrey, DE; Qiu, W; Chirgadze, NY; Mason, JM; Wei, X; Lin, DC; Che, Y; Kiarash, R; Fletcher, GC; Mak, TW; Bray, MR; Pauls, HW Discovery of Pyrazolo[1,5-a]pyrimidine TTK Inhibitors: CFI-402257 is a Potent, Selective, Bioavailable Anticancer Agent. ACS Med Chem Lett7:671-5 (2016) [PubMed] Article Liu, Y; Laufer, R; Patel, NK; Ng, G; Sampson, PB; Li, SW; Lang, Y; Feher, M; Brokx, R; Beletskaya, I; Hodgson, R; Plotnikova, O; Awrey, DE; Qiu, W; Chirgadze, NY; Mason, JM; Wei, X; Lin, DC; Che, Y; Kiarash, R; Fletcher, GC; Mak, TW; Bray, MR; Pauls, HW Discovery of Pyrazolo[1,5-a]pyrimidine TTK Inhibitors: CFI-402257 is a Potent, Selective, Bioavailable Anticancer Agent. ACS Med Chem Lett7:671-5 (2016) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 3A4 | |||
| Name: | Cytochrome P450 3A4 | ||
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 57349.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50512456 | |||
| n/a | |||
| Name | BDBM50512456 | ||
| Synonyms: | CHEMBL4469414 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H30N6O3 | ||
| Mol. Mass. | 498.5762 | ||
| SMILES | Cc1cc(ccc1C(=O)NC1CC1)-c1cnn2c(NC[C@H]3C[C@@](C)(O)C3)cc(Oc3cccnc3)nc12 |r,wU:20.21,22.25,(48.46,-32.37,;49.53,-31.26,;49.1,-29.78,;50.17,-28.67,;51.66,-29.03,;52.09,-30.5,;51.03,-31.63,;51.46,-33.1,;50.39,-34.22,;52.95,-33.47,;54.02,-32.36,;55.49,-31.92,;54.38,-30.85,;49.74,-27.2,;50.69,-25.98,;49.82,-24.7,;48.34,-25.14,;47.04,-24.33,;47.09,-22.79,;45.78,-21.98,;44.42,-22.71,;43.98,-24.18,;42.51,-23.74,;41.01,-23.33,;41.41,-24.82,;42.95,-22.26,;45.69,-25.06,;45.64,-26.6,;44.29,-27.33,;44.25,-28.87,;42.89,-29.59,;42.85,-31.13,;44.16,-31.94,;45.52,-31.2,;45.56,-29.66,;46.95,-27.4,;48.29,-26.68,)| | ||
| Structure | 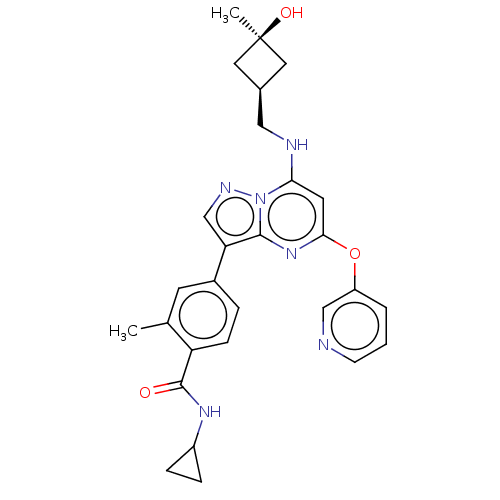
| ||